Significant Genes Associated with Mortality and Disease Progression in Grade II and III Glioma
- PMID: 38672212
- PMCID: PMC11048596
- DOI: 10.3390/biomedicines12040858
Significant Genes Associated with Mortality and Disease Progression in Grade II and III Glioma
Abstract
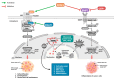
Background: The Wnt/β-catenin pathway plays a critical role in the tumorigenesis and maintenance of glioma stem cells. This study aimed to evaluate significant genes associated with the Wnt/β-catenin pathway involved in mortality and disease progression in patients with grade II and III glioma, using the Cancer Genome Atlas (TCGA) database.
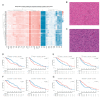
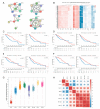
Methods: We obtained clinicopathological information and mRNA expression data from 515 patients with grade II and III gliomas from the TCGA database. We performed a multivariate Cox regression analysis to identify genes independently associated with glioma prognosis.
Results: The analysis of 34 genes involved in Wnt/β-catenin signaling demonstrated that four genes (CER1, FRAT1, FSTL1, and RPSA) related to the Wnt/β-catenin pathway were significantly associated with mortality and disease progression in patients with grade II and III glioma. We also identified additional genes related to the four significant genes of the Wnt/β-catenin pathway mentioned above. The higher expression of BMP2, RPL18A, RPL19, and RPS12 is associated with better outcomes in patients with glioma.
Conclusions: Using a large-scale open database, we identified significant genes related to the Wnt/β-catenin signaling pathway associated with mortality and disease progression in patients with grade II and III gliomas.
Keywords: TCGA; Wnt/β-catenin signaling; gene; glioma; survival.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures


References
-
- Mesfin F.B., Al-Dhahir M.A. StatPearls. StatPearls Publishing; Treasure Island, FL, USA: 2022. Gliomas.
-
- Miranda A., Hamilton P.T., Zhang A.W., Pattnaik S., Becht E., Mezheyeuski A., Bruun J., Micke P., de Reynies A., Nelson B.H. Cancer Stemness, Intratumoral Heterogeneity, and Immune Response across Cancers. Proc. Natl. Acad. Sci. USA. 2019;116:9020–9029. doi: 10.1073/pnas.1818210116. - DOI - PMC - PubMed
-
- Denysenko T., Annovazzi L., Cassoni P., Melcarne A., Mellai M., Schiffer D. WNT/β-Catenin Signaling Pathway and Downstream Modulators in Low- and High-Grade Glioma. Cancer Genom. Proteom. 2016;13:31–45. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

