Decoding Immuno-Competence: A Novel Analysis of Complete Blood Cell Count Data in COVID-19 Outcomes
- PMID: 38672225
- PMCID: PMC11048687
- DOI: 10.3390/biomedicines12040871
Decoding Immuno-Competence: A Novel Analysis of Complete Blood Cell Count Data in COVID-19 Outcomes
Abstract
Background: While 'immuno-competence' is a well-known term, it lacks an operational definition. To address this omission, this study explored whether the temporal and structured data of the complete blood cell count (CBC) can rapidly estimate immuno-competence. To this end, one or more ratios that included data on all monocytes, lymphocytes and neutrophils were investigated.
Materials and methods: Longitudinal CBC data collected from 101 COVID-19 patients (291 observations) were analyzed. Dynamics were estimated with several approaches, which included non-structured (the classic CBC format) and structured data. Structured data were assessed as complex ratios that capture multicellular interactions among leukocytes. In comparing survivors with non-survivors, the hypothesis that immuno-competence may exhibit feedback-like (oscillatory or cyclic) responses was tested.
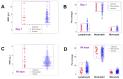
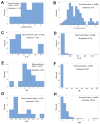
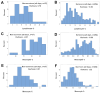
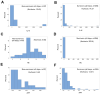
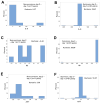
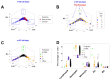
Results: While non-structured data did not distinguish survivors from non-survivors, structured data revealed immunological and statistical differences between outcomes: while survivors exhibited oscillatory data patterns, non-survivors did not. In survivors, many variables (including IL-6, hemoglobin and several complex indicators) showed values above or below the levels observed on day 1 of the hospitalization period, displaying L-shaped data distributions (positive kurtosis). In contrast, non-survivors did not exhibit kurtosis. Three immunologically defined data subsets included only survivors. Because information was based on visual patterns generated in real time, this method can, potentially, provide information rapidly.
Discussion: The hypothesis that immuno-competence expresses feedback-like loops when immunological data are structured was not rejected. This function seemed to be impaired in immuno-suppressed individuals. While this method rapidly informs, it is only a guide that, to be confirmed, requires additional tests. Despite this limitation, the fact that three protective (survival-associated) immunological data subsets were observed since day 1 supports many clinical decisions, including the early and personalized prognosis and identification of targets that immunomodulatory therapies could pursue. Because it extracts more information from the same data, structured data may replace the century-old format of the CBC.
Keywords: COVID-19; immune-competence; infectious disease.
Conflict of interest statement
A.L.R. and A.L.H. are co-inventors of the software utilized. All other authors declare no conflict of interest.
Figures






Similar articles
-
Personalized, disease-stage specific, rapid identification of immunosuppression in sepsis.Front Immunol. 2024 Oct 29;15:1430972. doi: 10.3389/fimmu.2024.1430972. eCollection 2024. Front Immunol. 2024. PMID: 39539549 Free PMC article.
-
COVID-19 disease-Temporal analyses of complete blood count parameters over course of illness, and relationship to patient demographics and management outcomes in survivors and non-survivors: A longitudinal descriptive cohort study.PLoS One. 2020 Dec 28;15(12):e0244129. doi: 10.1371/journal.pone.0244129. eCollection 2020. PLoS One. 2020. PMID: 33370366 Free PMC article.
-
Multi-Cellular Immunological Interactions Associated With COVID-19 Infections.Front Immunol. 2022 Feb 24;13:794006. doi: 10.3389/fimmu.2022.794006. eCollection 2022. Front Immunol. 2022. PMID: 35281033 Free PMC article.
-
Safety and efficacy assessment of allogeneic human dental pulp stem cells to treat patients with severe COVID-19: structured summary of a study protocol for a randomized controlled trial (Phase I / II).Trials. 2020 Jun 12;21(1):520. doi: 10.1186/s13063-020-04380-5. Trials. 2020. PMID: 32532356 Free PMC article.
-
The role of cytokine profile and lymphocyte subsets in the severity of coronavirus disease 2019 (COVID-19): A systematic review and meta-analysis.Life Sci. 2020 Oct 1;258:118167. doi: 10.1016/j.lfs.2020.118167. Epub 2020 Jul 29. Life Sci. 2020. PMID: 32735885 Free PMC article.
Cited by
-
Dynamics of Innate Immunity in SARS-CoV-2 Infections: Exploring the Impact of Natural Killer Cells, Inflammatory Responses, Viral Evasion Strategies, and Severity.Cells. 2025 May 22;14(11):763. doi: 10.3390/cells14110763. Cells. 2025. PMID: 40497938 Free PMC article. Review.
-
Personalized, disease-stage specific, rapid identification of immunosuppression in sepsis.Front Immunol. 2024 Oct 29;15:1430972. doi: 10.3389/fimmu.2024.1430972. eCollection 2024. Front Immunol. 2024. PMID: 39539549 Free PMC article.
References
-
- Grimm C., Dickel S., Grundmann J., Payen D., Schanz J., Zautner A.E., Tampe B., Moerer O., Winkler M.S. Case Report: Interferon-g Rescues Monocytic Human Leukocyte Antigen Receptor (mHLA-DR) Function in a COVID-19 Patient With ARDS and Superinfection With Multiple MDR 4MRGN Bacterial Strains. Front. Immunol. 2021;12:753849. doi: 10.3389/fimmu.2021.753849. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources

