Comprehensive Multi-Omic Evaluation of the Microbiota and Metabolites in the Colons of Diverse Swine Breeds
- PMID: 38672368
- PMCID: PMC11047667
- DOI: 10.3390/ani14081221
Comprehensive Multi-Omic Evaluation of the Microbiota and Metabolites in the Colons of Diverse Swine Breeds
Abstract
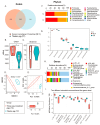
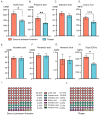
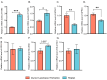
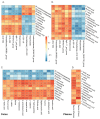
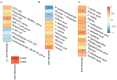
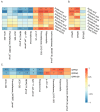
Pigs stand as a vital cornerstone in the realm of human sustenance, and the intricate composition of their intestinal microbiota wields a commanding influence over their nutritional and metabolic pathways. We employed multi-omic evaluations to identify microbial evidence associated with differential growth performance and metabolites, thereby offering theoretical support for the implementation of efficient farming practices for Tibetan pigs and establishing a robust foundation for enhancing pig growth and health. In this work, six Duroc × landrace × yorkshi (DLY) pigs and six Tibetan pigs were used for the experiment. Following humane euthanasia, a comprehensive analysis was undertaken to detect the presence of short-chain fatty acids (SCFAs), microbial populations, and metabolites within the colonic environment. Additionally, metabolites present within the plasma were also assessed. The outcomes of our analysis unveiled the key variables affecting the microbe changes causing the observed differences in production performance between these two distinct pig breeds. Specifically, noteworthy discrepancies were observed in the microbial compositions of DLY pigs, characterized by markedly higher levels of Alloprevotella and Prevotellaceae_UCG-003 (p < 0.05). These disparities, in turn, resulted in significant variations in the concentrations of acetic acid, propionic acid, and the cumulative SCFAs (p < 0.05). Consequently, the DLY pigs exhibited enhanced growth performance and overall well-being, which could be ascribed to the distinct metabolite profiles they harbored. Conversely, Tibetan pigs exhibited a significantly elevated relative abundance of the NK4A214_group, which consequently led to a pronounced increase in the concentration of L-cysteine. This elevation in L-cysteine content had cascading effects, further manifesting higher levels of taurine within the colon and plasma. It is noteworthy that taurine has the potential to exert multifaceted impacts encompassing microbiota dynamics, protein and lipid metabolism, as well as bile acid metabolism, all of which collectively benefit the pigs. In light of this, Tibetan pigs showcased enhanced capabilities in bile acid metabolism. In summation, our findings suggest that DLY pigs excel in their proficiency in short-chain fatty acid metabolism, whereas Tibetan pigs exhibit a more pronounced competence in the realm of bile acid metabolism. These insights underscore the potential for future studies to leverage these breed-specific differences, thereby contributing to the amelioration of production performance within these two distinct pig breeds.
Keywords: SCFA; gut; microbiota; nutrition; pigs.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
Multiomics comparative analysis of feces AMRGs of Duroc pigs and Tibetan and the effect of fecal microbiota transplantation on AMRGs upon antibiotic exposure.Microbiol Spectr. 2024 Nov 29;13(5):e0198324. doi: 10.1128/spectrum.01983-24. Online ahead of print. Microbiol Spectr. 2024. PMID: 39612216 Free PMC article.
-
Comparative Study on Jejunal Immunity and Microbial Composition of Growing-Period Tibetan Pigs and Duroc × (Landrace × Yorkshire) Pigs.Front Vet Sci. 2022 Apr 25;9:890585. doi: 10.3389/fvets.2022.890585. eCollection 2022. Front Vet Sci. 2022. PMID: 35548051 Free PMC article.
-
Insights into high-altitude adaptation and meat quality regulation by gastrointestinal metabolites in Tibetan and black pigs.Front Vet Sci. 2025 Mar 26;12:1569196. doi: 10.3389/fvets.2025.1569196. eCollection 2025. Front Vet Sci. 2025. PMID: 40206253 Free PMC article.
-
Enhanced immunity: the gut microbiota changes in high-altitude Tibetan pigs compared to Yorkshire pigs.Front Microbiol. 2024 Nov 18;15:1469253. doi: 10.3389/fmicb.2024.1469253. eCollection 2024. Front Microbiol. 2024. PMID: 39624721 Free PMC article.
-
The interaction between dietary fiber and gut microbiota, and its effect on pig intestinal health.Front Immunol. 2023 Feb 14;14:1095740. doi: 10.3389/fimmu.2023.1095740. eCollection 2023. Front Immunol. 2023. PMID: 36865557 Free PMC article. Review.
Cited by
-
Identification and Functional Validation of ACSL1 and FABP3 as Muscle-Related Genes Screened by Transcriptomics in Crossbred Duroc × Berkshire × Diannan Small-Eared Pigs.Genes (Basel). 2025 Apr 29;16(5):520. doi: 10.3390/genes16050520. Genes (Basel). 2025. PMID: 40428342 Free PMC article.
References
Grants and funding
LinkOut - more resources
Full Text Sources

