Oropharyngeal Microbiome Analysis in Patients with Varying SARS-CoV-2 Infection Severity: A Prospective Cohort Study
- PMID: 38672996
- PMCID: PMC11051038
- DOI: 10.3390/jpm14040369
Oropharyngeal Microbiome Analysis in Patients with Varying SARS-CoV-2 Infection Severity: A Prospective Cohort Study
Abstract
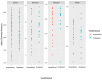
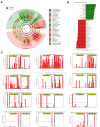
Patients with COVID-19 infection have distinct oropharyngeal microbiota composition and diversity metrics according to disease severity. However, these findings are not consistent across the literature. We conducted a multicenter, prospective study in patients with COVID-19 requiring outpatient versus inpatient management to explore the microbial abundance of taxa at the phylum, family, genus, and species level, and we utilized alpha and beta diversity indices to further describe our findings. We collected oropharyngeal washing specimens at the time of study entry, which coincided with the COVID-19 diagnosis, to conduct all analyses. We included 43 patients in the study, of whom 16 were managed as outpatients and 27 required hospitalization. Proteobacteria, Actinobacteria, Bacteroidetes, Saccharibacteria TM7, Fusobacteria, and Spirochaetes were the most abundant phyla among patients, while 61 different families were detected, of which the Streptococcaceae and Staphylococcaceae families were the most predominant. A total of 132 microbial genera were detected, with Streptococcus being the predominant genus in outpatients, in contrast to hospitalized patients, in whom the Staphylococcus genus was predominant. LeFSe analysis identified 57 microbial species in the oropharyngeal washings of study participants that could discriminate the severity of symptoms of COVID-19 infections. Alpha diversity analysis did not reveal a difference in the abundance of bacterial species between the groups, but beta diversity analysis established distinct microbial communities between inpatients and outpatients. Our study provides information on the complex association between the oropharyngeal microbiota and SARS-CoV-2 infection. Although our study cannot establish causation, knowledge of specific taxonomic changes with increasing SARS-CoV-2 infection severity can provide us with novel clues for the prognostic classification of COVID-19 patients.
Keywords: SARS-CoV-2 infection; bacterial abundance; bacterial diversity; oropharyngeal microbiome.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures



References
-
- Kaeuffer C., Le Hyaric C., Fabacher T., Mootien J., Dervieux B., Ruch Y., Hugerot A., Zhu Y.J., Pointurier V., Clere-Jehl R., et al. Clinical characteristics and risk factors associated with severe COVID-19: Prospective analysis of 1045 hospitalised cases in North-Eastern France, March 2020. Euro Surveill. 2020;25:2000895. doi: 10.2807/1560-7917.ES.2020.25.48.2000895. - DOI - PMC - PubMed
-
- Milota T., Sobotkova M., Smetanova J., Bloomfield M., Vydlakova J., Chovancova Z., Litzman J., Hakl R., Novak J., Malkusova I., et al. Risk Factors for Severe COVID-19 and Hospital Admission in Patients with Inborn Errors of Immunity—Results from a Multicenter Nationwide Study. Front. Immunol. 2022;13:835770. doi: 10.3389/fimmu.2022.835770. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

