Computational Analysis of Deleterious nsSNPs in INS Gene Associated with Permanent Neonatal Diabetes Mellitus
- PMID: 38673052
- PMCID: PMC11051494
- DOI: 10.3390/jpm14040425
Computational Analysis of Deleterious nsSNPs in INS Gene Associated with Permanent Neonatal Diabetes Mellitus
Abstract
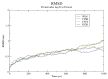
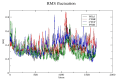
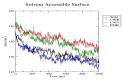
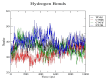
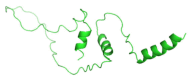
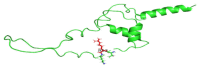
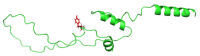
Insulin gene mutations affect the structure of insulin and are considered a leading cause of neonatal diabetes and permanent neonatal diabetes mellitus PNDM. These mutations can affect the production and secretion of insulin, resulting in inadequate insulin levels and subsequent hyperglycemia. Early discovery or prediction of PNDM can aid in better management and treatment. The current study identified potential deleterious non-synonymous single nucleotide polymorphisms nsSNPs in the INS gene. The analysis of the nsSNPs in the INS gene was conducted using bioinformatics tools by implementing computational algorithms including SIFT, PolyPhen2, SNAP2, SNPs & GO, PhD-SNP, MutPred2, I-Mutant, MuPro, and HOPE tools to investigate the prediction of the potential association between nsSNPs in the INS gene and PNDM. Three mutations, C96Y, P52R, and C96R, were shown to potentially reduce the stability and function of the INS protein. These mutants were subjected to MDSs for structural analysis. Results suggested that these three potential pathogenic mutations may affect the stability and functionality of the insulin protein encoded by the INS gene. Therefore, these changes may influence the development of PNDM. Further researches are required to fully understand the various effects of mutations in the INS gene on insulin synthesis and function. These data can aid in genetic testing for PNDM to evaluate its risk and create treatment and prevention strategies in personalized medicine.
Keywords: computational methods; genetic variations; insulin genes; non-synonymous single nucleotide polymorphisms; permanent neonatal diabetes mellitus; personalized medicine.
Conflict of interest statement
The authors declare that no conflicts of interest exist.
Figures



References
-
- Wang H., Saint-Martin C., Xu J., Ding L., Wang R., Feng W., Liu M., Shu H., Fan Z., Haataja L., et al. Biological behaviors of mutant proinsulin contribute to the phenotypic spectrum of diabetes associated with insulin gene mutations. Mol. Cell. Endocrinol. 2020;518:111025. doi: 10.1016/j.mce.2020.111025. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources

