Transcriptome Profiling Unveils Key Genes Regulating the Growth and Development of Yangzhou Goose Knob
- PMID: 38673752
- PMCID: PMC11050116
- DOI: 10.3390/ijms25084166
Transcriptome Profiling Unveils Key Genes Regulating the Growth and Development of Yangzhou Goose Knob
Abstract
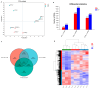
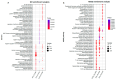
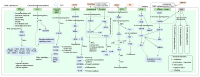
Goose is one of the most economically valuable poultry species and has a distinct appearance due to its possession of a knob. A knob is a hallmark of sexual maturity in goose (Anser cygnoides) and plays crucial roles in artificial selection, health status, social signaling, and body temperature regulation. However, the genetic mechanisms influencing the growth and development of goose knobs remain completely unclear. In this study, histomorphological and transcriptomic analyses of goose knobs in D70, D120, and D300 Yangzhou geese revealed differential changes in tissue morphology during the growth and development of goose knobs and the key core genes that regulate goose knob traits. Observation of tissue sections revealed that as age increased, the thickness of the knob epidermis, cuticle, and spinous cells gradually decreased. Additionally, fat cells in the dermis and subcutaneous connective tissue transitioned from loose to dense. Transcriptome sequencing results, analyzed through differential expression, Weighted Gene Co-expression Network Analysis (WGCNA), and pattern expression analysis methods, showed D70-vs.-D120 (up-regulated: 192; down-regulated: 423), D70-vs.-D300 (up-regulated: 1394; down-regulated: 1893), and D120-vs.-D300 (up-regulated: 1017; down-regulated: 1324). A total of 6243 differentially expressed genes (DEGs) were identified, indicating varied expression levels across the three groups in the knob tissues of D70, D120, and D300 Yangzhou geese. These DEGs are significantly enriched in biological processes (BP) such as skin morphogenesis, the regulation of keratinocyte proliferation, and epidermal cell differentiation. Furthermore, they demonstrate enrichment in pathways related to goose knob development, including ECM-receptor interaction, NF-kappa B, and PPAR signaling. Through pattern expression analysis, three gene expression clusters related to goose knob traits were identified. The joint analysis of candidate genes associated with goose knob development and WGCNA led to the identification of key core genes influencing goose knob development. These core genes comprise WNT4, WNT10A, TCF7L2, GATA3, ADRA2A, CASP3, SFN, KDF1, ERRFI1, SPRY1, and EVPL. In summary, this study provides a reference for understanding the molecular mechanisms of goose knob growth and development and provides effective ideas and methods for the genetic improvement of goose knob traits.
Keywords: DEGs; RNA-seq; WGCNA; Yangzhou goose; knob.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures






References
-
- FAO-STAT Food and Agriculture Organization of the United Nations. Livestock Primary. 2023. [(accessed on 10 July 2023)]. Available online: https://www.fao.org/faostat/en/#data/QL.
-
- Wright D., Rubin C., Schutz K., Kerje S., Kindmark A., Brandström H., Andersson L., Pizzari T., Jensen P. Onset of sexual maturity in female chickens is genetically linked to loci associated with fecundity and a sexual ornament. Reprod. Domest. Anim. = Zuchthygiene. 2012;47((Suppl. 1)):31–36. doi: 10.1111/j.1439-0531.2011.01963.x. - DOI - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

