Entamoeba histolytica: In Silico and In Vitro Oligomerization of EhHSTF5 Enhances Its Binding to the HSE of the EhPgp5 Gene Promoter
- PMID: 38673804
- PMCID: PMC11050682
- DOI: 10.3390/ijms25084218
Entamoeba histolytica: In Silico and In Vitro Oligomerization of EhHSTF5 Enhances Its Binding to the HSE of the EhPgp5 Gene Promoter
Abstract
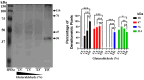
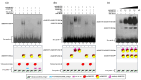
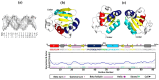
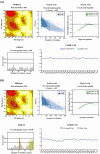
Throughout its lifecycle, Entamoeba histolytica encounters a variety of stressful conditions. This parasite possesses Heat Shock Response Elements (HSEs) which are crucial for regulating the expression of various genes, aiding in its adaptation and survival. These HSEs are regulated by Heat Shock Transcription Factors (EhHSTFs). Our research has identified seven such factors in the parasite, designated as EhHSTF1 through to EhHSTF7. Significantly, under heat shock conditions and in the presence of the antiamoebic compound emetine, EhHSTF5, EhHSTF6, and EhHSTF7 show overexpression, highlighting their essential role in gene response to these stressors. Currently, only EhHSTF7 has been confirmed to recognize the HSE as a promoter of the EhPgp5 gene (HSE_EhPgp5), leaving the binding potential of the other EhHSTFs to HSEs yet to be explored. Consequently, our study aimed to examine, both in vitro and in silico, the oligomerization, and binding capabilities of the recombinant EhHSTF5 protein (rEhHSTF5) to HSE_EhPgp5. The in vitro results indicate that the oligomerization of rEhHSTF5 is concentration-dependent, with its dimeric conformation showing a higher affinity for HSE_EhPgp5 than its monomeric state. In silico analysis suggests that the alpha 3 α-helix (α3-helix) of the DNA-binding domain (DBD5) of EhHSTF5 is crucial in binding to the major groove of HSE, primarily through hydrogen bonding and salt-bridge interactions. In summary, our results highlight the importance of oligomerization in enhancing the affinity of rEhHSTF5 for HSE_EhPgp5 and demonstrate its ability to specifically recognize structural motifs within HSE_EhPgp5. These insights significantly contribute to our understanding of one of the potential molecular mechanisms employed by this parasite to efficiently respond to various stressors, thereby enabling successful adaptation and survival within its host environment.
Keywords: EhDBD5; Entamoeba histolytica; HSE_EhPgp5; dimer; molecular docking; monomer; oligomerization; rEhHSTF5; trimer.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures




Similar articles
-
Screening and Structural Characterization of Heat Shock Response Elements (HSEs) in Entamoeba histolytica Promoters.Int J Mol Sci. 2024 Jan 21;25(2):1319. doi: 10.3390/ijms25021319. Int J Mol Sci. 2024. PMID: 38279319 Free PMC article.
-
A Novel Heat Shock Element (HSE) in Entamoeba histolytica that Regulates the Transcriptional Activation of the EhPgp5 Gene in the Presence of Emetine Drug.Front Cell Infect Microbiol. 2017 Nov 29;7:492. doi: 10.3389/fcimb.2017.00492. eCollection 2017. Front Cell Infect Microbiol. 2017. PMID: 29238701 Free PMC article.
-
The novel EhHSTF7 transcription factor displays an oligomer state and recognizes a heat shock element in the Entamoeba histolytica parasite.Microb Pathog. 2022 Jan;162:105349. doi: 10.1016/j.micpath.2021.105349. Epub 2021 Dec 2. Microb Pathog. 2022. PMID: 34864144
-
Multidrug resistance in the protozoan parasite Entamoeba histolytica.Parasitol Int. 2002 Dec;51(4):353-9. doi: 10.1016/s1383-5769(02)00041-7. Parasitol Int. 2002. PMID: 12421633 Review.
-
Physiology and molecular biology of multidrug resistance in Entamoeba histolytica.Arch Med Res. 1996 Autumn;27(3):421-5. Arch Med Res. 1996. PMID: 8854404 Review.
Cited by
-
Polyphenols from Bacopa procumbens Nanostructured with Gold Nanoparticles Stimulate Hair Growth Through Apoptosis Modulation in C57BL/6 Mice.Pharmaceutics. 2025 Feb 9;17(2):222. doi: 10.3390/pharmaceutics17020222. Pharmaceutics. 2025. PMID: 40006589 Free PMC article.
References
-
- Chou A., Austin R.L. StatPearls. StatPearls Publishing; Treasure Island, FL, USA: 2023. Entamoeba Histolytica Infection. - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

