Duocarmycin SA Reduces Proliferation and Increases Apoptosis in Acute Myeloid Leukemia Cells In Vitro
- PMID: 38673926
- PMCID: PMC11050052
- DOI: 10.3390/ijms25084342
Duocarmycin SA Reduces Proliferation and Increases Apoptosis in Acute Myeloid Leukemia Cells In Vitro
Abstract
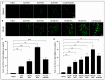
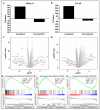
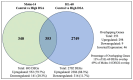
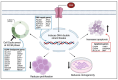
Acute myeloid leukemia (AML) is a hematological malignancy that is characterized by an expansion of immature myeloid precursors. Despite therapeutic advances, the prognosis of AML patients remains poor and there is a need for the evaluation of promising therapeutic candidates to treat the disease. The objective of this study was to evaluate the efficacy of duocarmycin Stable A (DSA) in AML cells in vitro. We hypothesized that DSA would induce DNA damage in the form of DNA double-strand breaks (DSBs) and exert cytotoxic effects on AML cells within the picomolar range. Human AML cell lines Molm-14 and HL-60 were used to perform 3-(4,5-dimethylthiazolyl-2)-2,5-diphenyltetrazolium bromide (MTT), DNA DSBs, cell cycle, 5-ethynyl-2-deoxyuridine (EdU), colony formation unit (CFU), Annexin V, RNA sequencing and other assays described in this study. Our results showed that DSA induced DNA DSBs, induced cell cycle arrest at the G2M phase, reduced proliferation and increased apoptosis in AML cells. Additionally, RNA sequencing results showed that DSA regulates genes that are associated with cellular processes such as DNA repair, G2M checkpoint and apoptosis. These results suggest that DSA is efficacious in AML cells and is therefore a promising potential therapeutic candidate that can be further evaluated for the treatment of AML.
Keywords: DNA alkylation; DNA double-strand break; acute myeloid leukemia; apoptosis; chemotherapy; duocarmycin SA.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures






Similar articles
-
Tetrahydroindazolone-substituted Benzamide Compound W-H4 Induces Apoptosis and Autophagy of Acute Myeloid Leukemia Cells.Anticancer Res. 2025 Jul;45(7):2985-2996. doi: 10.21873/anticanres.17664. Anticancer Res. 2025. PMID: 40578939
-
Interleukin-2 as maintenance therapy for children and adults with acute myeloid leukaemia in first complete remission.Cochrane Database Syst Rev. 2015 Nov 6;2015(11):CD010248. doi: 10.1002/14651858.CD010248.pub2. Cochrane Database Syst Rev. 2015. PMID: 26544114 Free PMC article.
-
Thrombopoietin mimetics for patients with myelodysplastic syndromes.Cochrane Database Syst Rev. 2017 Sep 30;9(9):CD009883. doi: 10.1002/14651858.CD009883.pub2. Cochrane Database Syst Rev. 2017. PMID: 28962071 Free PMC article.
-
Comparison of first-line chemotherapy including escalated BEACOPP versus chemotherapy including ABVD for people with early unfavourable or advanced stage Hodgkin lymphoma.Cochrane Database Syst Rev. 2017 May 25;5(5):CD007941. doi: 10.1002/14651858.CD007941.pub3. Cochrane Database Syst Rev. 2017. PMID: 28541603 Free PMC article.
-
A rapid and systematic review of the clinical effectiveness and cost-effectiveness of paclitaxel, docetaxel, gemcitabine and vinorelbine in non-small-cell lung cancer.Health Technol Assess. 2001;5(32):1-195. doi: 10.3310/hta5320. Health Technol Assess. 2001. PMID: 12065068
Cited by
-
Prodrugs in Oncology: Bioactivation and Impact on Therapeutic Efficacy and Toxicity.Int J Mol Sci. 2025 Jan 24;26(3):988. doi: 10.3390/ijms26030988. Int J Mol Sci. 2025. PMID: 39940757 Free PMC article. Review.
-
A Comprehensive Review of the Antitumor Properties and Mechanistic Insights of Duocarmycin Analogs.Cancers (Basel). 2024 Sep 27;16(19):3293. doi: 10.3390/cancers16193293. Cancers (Basel). 2024. PMID: 39409913 Free PMC article. Review.
References
-
- Howlader N., Noone A., Krapcho M., Miller D., Brest A., Yu M., Ruhl J., Tatalovich Z., Mariotto A., Lewis D.R., et al. SEER Cancer Statistics Review, 1975–2017. National Cancer Institute; Bethesda, MD, USA: 2020.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

