PfbZIP85 Transcription Factor Mediates ω-3 Fatty Acid-Enriched Oil Biosynthesis by Down-Regulating PfLPAT1B Gene Expression in Plant Tissues
- PMID: 38673960
- PMCID: PMC11050522
- DOI: 10.3390/ijms25084375
PfbZIP85 Transcription Factor Mediates ω-3 Fatty Acid-Enriched Oil Biosynthesis by Down-Regulating PfLPAT1B Gene Expression in Plant Tissues
Abstract
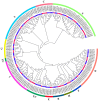
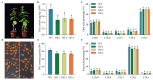
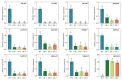
The basic leucine zipper (bZIP) transcription factor (TF) family is one of the biggest TF families identified so far in the plant kingdom, functioning in diverse biological processes including plant growth and development, signal transduction, and stress responses. For Perilla frutescens, a novel oilseed crop abundant in polyunsaturated fatty acids (PUFAs) (especially α-linolenic acid, ALA), the identification and biological functions of bZIP members remain limited. In this study, 101 PfbZIPs were identified in the perilla genome and classified into eleven distinct groups (Groups A, B, C, D, E, F, G, H, I, S, and UC) based on their phylogenetic relationships and gene structures. These PfbZIP genes were distributed unevenly across 18 chromosomes, with 83 pairs of them being segmental duplication genes. Moreover, 78 and 148 pairs of orthologous bZIP genes were detected between perilla and Arabidopsis or sesame, respectively. PfbZIP members belonging to the same subgroup exhibited highly conserved gene structures and functional domains, although significant differences were detected between groups. RNA-seq and RT-qPCR analysis revealed differential expressions of 101 PfbZIP genes during perilla seed development, with several PfbZIPs exhibiting significant correlations with the key oil-related genes. Y1H and GUS activity assays evidenced that PfbZIP85 downregulated the expression of the PfLPAT1B gene by physical interaction with the promoter. PfLPAT1B encodes a lysophosphatidate acyltransferase (LPAT), one of the key enzymes for triacylglycerol (TAG) assembly. Heterogeneous expression of PfbZIP85 significantly reduced the levels of TAG and UFAs (mainly C18:1 and C18:2) but enhanced C18:3 accumulation in both seeds and non-seed tissues in the transgenic tobacco lines. Furthermore, these transgenic tobacco plants showed no significantly adverse phenotype for other agronomic traits such as plant growth, thousand seed weight, and seed germination rate. Collectively, these findings offer valuable perspectives for understanding the functions of PfbZIPs in perilla, particularly in lipid metabolism, showing PfbZIP85 as a suitable target in plant genetic improvement for high-value vegetable oil production.
Keywords: bZIP transcription factors; expression analysis; genome-wide identification; lysophosphatidate acyltransferase (LPAT); oil biosynthesis and regulation; perilla (Perilla frutescens).
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial commitments that could be construed as a potential conflict of interest.
Figures







Similar articles
-
Genome-Wide Analysis of Glycerol-3-Phosphate Acyltransferase (GPAT) Family in Perilla frutescens and Functional Characterization of PfGPAT9 Crucial for Biosynthesis of Storage Oils Rich in High-Value Lipids.Int J Mol Sci. 2023 Oct 12;24(20):15106. doi: 10.3390/ijms242015106. Int J Mol Sci. 2023. PMID: 37894786 Free PMC article.
-
[Cloning and functional characterization of a lysophosphatidic acid acyltransferase gene from Perilla frutescens].Sheng Wu Gong Cheng Xue Bao. 2022 Aug 25;38(8):3014-3028. doi: 10.13345/j.cjb.22003. Sheng Wu Gong Cheng Xue Bao. 2022. PMID: 36002428 Chinese.
-
Genome-wide analysis of DGAT gene family in Perilla frutescens and functional characterization of PfDGAT2-2 and PfDGAT3-1 in Arabidopsis.Plant Sci. 2022 Nov;324:111426. doi: 10.1016/j.plantsci.2022.111426. Epub 2022 Aug 23. Plant Sci. 2022. PMID: 35998725
-
Regulation of Oil Biosynthesis and Genetic Improvement in Plants: Advances and Prospects.Genes (Basel). 2024 Aug 26;15(9):1125. doi: 10.3390/genes15091125. Genes (Basel). 2024. PMID: 39336716 Free PMC article. Review.
-
Role of bZIP Transcription Factors in Plant Salt Stress.Int J Mol Sci. 2023 Apr 26;24(9):7893. doi: 10.3390/ijms24097893. Int J Mol Sci. 2023. PMID: 37175598 Free PMC article. Review.
Cited by
-
Genome-wide profiling of bZIP transcription factors in Camelina sativa: implications for development and stress response.BMC Genom Data. 2024 Oct 14;25(1):88. doi: 10.1186/s12863-024-01270-6. BMC Genom Data. 2024. PMID: 39402491 Free PMC article.
-
Advancements in Crop PUFAs Biosynthesis and Genetic Engineering: A Systematic and Mixed Review System.Int J Mol Sci. 2025 Apr 8;26(8):3462. doi: 10.3390/ijms26083462. Int J Mol Sci. 2025. PMID: 40331974 Free PMC article.
-
Identification and expression analysis of bZIP transcription factors in Setaria italica in response to dehydration stress.Front Genet. 2024 Aug 30;15:1466486. doi: 10.3389/fgene.2024.1466486. eCollection 2024. Front Genet. 2024. PMID: 39280094 Free PMC article.
References
-
- Nitta M., Lee J.K., Kang C.W., Katsuta M., Yasumoto S., Liu D., Nagamine T., Ohnishi O. The Distribution of Perilla Species. Genet. Resour. Crop Evol. 2005;52:797–804. doi: 10.1007/s10722-003-6017-5. - DOI
-
- Park Y.J., Dixit A., Ma K.H., Lee J.K., Lee M.H., Chung C.S., Nitta M., Okuno K., Kim T.S., Cho E.G., et al. Evaluation of genetic diversity and relationships within an on-farm collection of Perilla frutescens (L.) Britt. using microsatellite markers. Genet. Resour. Crop Evol. 2008;55:523–535. doi: 10.1007/s10722-007-9258-x. - DOI
MeSH terms
Grants and funding
- YZ2021-08/the Breeding Engineering Special Key Cultivation Project of Agricultural College, Shanxi Agricul-tural University
- 20210302123418/the Basic Research Program (Free Exploration) Project of Shanxi Province, China
- CXGC2023048/the Science and Technology Innovation Project, Shanxi Agricultural University
- 31201266/the National Natural Science Foundation of China
- 31401430/The National Natural Science Foundation of China
LinkOut - more resources
Full Text Sources
Miscellaneous

