The Inhibition of NS2B/NS3 Protease: A New Therapeutic Opportunity to Treat Dengue and Zika Virus Infection
- PMID: 38673962
- PMCID: PMC11050111
- DOI: 10.3390/ijms25084376
The Inhibition of NS2B/NS3 Protease: A New Therapeutic Opportunity to Treat Dengue and Zika Virus Infection
Abstract
In the global pandemic scenario, dengue and zika viruses (DENV and ZIKV, respectively), both mosquito-borne members of the flaviviridae family, represent a serious health problem, and considering the absence of specific antiviral drugs and available vaccines, there is a dire need to identify new targets to treat these types of viral infections. Within this drug discovery process, the protease NS2B/NS3 is considered the primary target for the development of novel anti-flavivirus drugs. The NS2B/NS3 is a serine protease that has a dual function both in the viral replication process and in the elusion of the innate immunity. To date, two main classes of NS2B/NS3 of DENV and ZIKV protease inhibitors have been discovered: those that bind to the orthosteric site and those that act at the allosteric site. Therefore, this perspective article aims to discuss the main features of the use of the most potent NS2B/NS3 inhibitors and their impact at the social level.
Keywords: NS2B/NS3 serine protease; antiviral agents; dengue virus; orthosteric and allosteric inhibitors; zika virus.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures

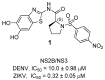
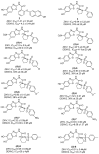

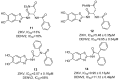
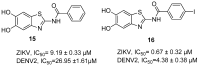
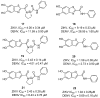
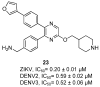
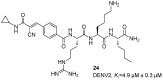


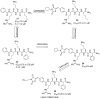
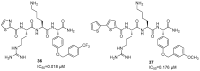
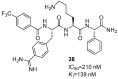
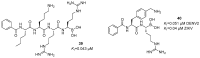
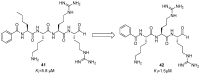





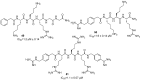

References
-
- World Health Organization Dengue and Severe Dengue. [(accessed on 15 December 2023)]; Available online: https://www.who.int/news-room/fact-sheets/detail/dengue-and-severe-dengue.
-
- World Health Organization Zika Virus. [(accessed on 15 December 2023)]; Available online: https://www.who.int/news-room/fact-sheets/detail/zika-virus.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

