Novel Concept of Alpha Satellite Cascading Higher-Order Repeats (HORs) and Precise Identification of 15mer and 20mer Cascading HORs in Complete T2T-CHM13 Assembly of Human Chromosome 15
- PMID: 38673983
- PMCID: PMC11050224
- DOI: 10.3390/ijms25084395
Novel Concept of Alpha Satellite Cascading Higher-Order Repeats (HORs) and Precise Identification of 15mer and 20mer Cascading HORs in Complete T2T-CHM13 Assembly of Human Chromosome 15
Abstract
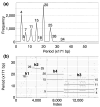
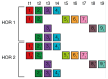
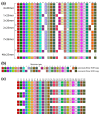
Unraveling the intricate centromere structure of human chromosomes holds profound implications, illuminating fundamental genetic mechanisms and potentially advancing our comprehension of genetic disorders and therapeutic interventions. This study rigorously identified and structurally analyzed alpha satellite higher-order repeats (HORs) within the centromere of human chromosome 15 in the complete T2T-CHM13 assembly using the high-precision GRM2023 algorithm. The most extensive alpha satellite HOR array in chromosome 15 reveals a novel cascading HOR, housing 429 15mer HOR copies, containing 4-, 7- and 11-monomer subfragments. Within each row of cascading HORs, all alpha satellite monomers are of distinct types, as in regular Willard's HORs. However, different HOR copies within the same cascading 15mer HOR contain more than one monomer of the same type. Each canonical 15mer HOR copy comprises 15 monomers belonging to only 9 different monomer types. Notably, 65% of the 429 15mer cascading HOR copies exhibit canonical structures, while 35% display variant configurations. Identified as the second most extensive alpha satellite HOR, another novel cascading HOR within human chromosome 15 encompasses 164 20mer HOR copies, each featuring two subfragments. Moreover, a distinct pattern emerges as interspersed 25mer/26mer structures differing from regular Willard's HORs and giving rise to a 34-monomer subfragment. Only a minor 18mer HOR array of 12 HOR copies is of the regular Willard's type. These revelations highlight the complexity within the chromosome 15 centromeric region, accentuating deviations from anticipated highly regular patterns and hinting at profound information encoding and functional potential within the human centromere.
Keywords: GRM algorithm; T2T-CHM13 human assembly; alpha satellites; centromere; higher-order repeats.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures





Similar articles
-
Novel Cascade Alpha Satellite HORs in Orangutan Chromosome 13 Assembly: Discovery of the 59mer HOR-The largest Unit in Primates-And the Missing Triplet 45/27/18 HOR in Human T2T-CHM13v2.0 Assembly.Int J Mol Sci. 2024 Jul 11;25(14):7596. doi: 10.3390/ijms25147596. Int J Mol Sci. 2024. PMID: 39062839 Free PMC article.
-
Precise identification of cascading alpha satellite higher order repeats in T2T-CHM13 assembly of human chromosome 3.Croat Med J. 2024 Jun 13;65(3):209-219. doi: 10.3325/cmj.2024.65.209. Croat Med J. 2024. PMID: 38868967 Free PMC article.
-
Global Repeat Map (GRM): Advantageous Method for Discovery of Largest Higher-Order Repeats (HORs) in Neuroblastoma Breakpoint Family (NBPF) Genes, in Hornerin Exon and in Chromosome 21 Centromere.Prog Mol Subcell Biol. 2021;60:203-234. doi: 10.1007/978-3-030-74889-0_8. Prog Mol Subcell Biol. 2021. PMID: 34386877
-
Key-string algorithm--novel approach to computational analysis of repetitive sequences in human centromeric DNA.Croat Med J. 2003 Aug;44(4):386-406. Croat Med J. 2003. PMID: 12950141 Review.
-
Dynamic interplay between human alpha-satellite DNA structure and centromere functions.Semin Cell Dev Biol. 2024 Mar 15;156:130-140. doi: 10.1016/j.semcdb.2023.10.002. Epub 2023 Nov 4. Semin Cell Dev Biol. 2024. PMID: 37926668 Review.
Cited by
-
Novel Cascade Alpha Satellite HORs in Orangutan Chromosome 13 Assembly: Discovery of the 59mer HOR-The largest Unit in Primates-And the Missing Triplet 45/27/18 HOR in Human T2T-CHM13v2.0 Assembly.Int J Mol Sci. 2024 Jul 11;25(14):7596. doi: 10.3390/ijms25147596. Int J Mol Sci. 2024. PMID: 39062839 Free PMC article.
References
-
- Miga K.H. The Promises and Challenges of Genomic Studies of Human Centromeres. Prog. Mol. Subcell Biol. 2017;56:285–304. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

