Moving toward the Inclusion of Epigenomics in Bacterial Genome Evolution: Perspectives and Challenges
- PMID: 38674013
- PMCID: PMC11050019
- DOI: 10.3390/ijms25084425
Moving toward the Inclusion of Epigenomics in Bacterial Genome Evolution: Perspectives and Challenges
Abstract
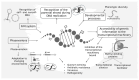
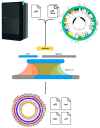
The universality of DNA methylation as an epigenetic regulatory mechanism belongs to all biological kingdoms. However, while eukaryotic systems have been the primary focus of DNA methylation studies, the molecular mechanisms in prokaryotes are less known. Nevertheless, DNA methylation in prokaryotes plays a pivotal role in many cellular processes such as defense systems against exogenous DNA, cell cycle dynamics, and gene expression, including virulence. Thanks to single-molecule DNA sequencing technologies, genome-wide identification of methylated DNA is becoming feasible on a large scale, providing the possibility to investigate more deeply the presence, variability, and roles of DNA methylation. Here, we present an overview of the multifaceted roles of DNA methylation in prokaryotes and suggest research directions and tools which can enable us to better understand the contribution of DNA methylation to prokaryotic genome evolution and adaptation. In particular, we emphasize the need to understand the presence and role of transgenerational inheritance, as well as the impact of epigenomic signatures on adaptation and genome evolution. Research directions and the importance of novel computational tools are underlined.
Keywords: bacterial epigenomics; bacterial genome evolution; prokaryotic DNA methylation.
Conflict of interest statement
The authors declare no conflicts of interest. The funders had no role in the design of the study, nor in the collection, analyses, or interpretation of the data.
Figures
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

