Genomic and Phenotypic Analysis of Salmonella enterica Bacteriophages Identifies Two Novel Phage Species
- PMID: 38674639
- PMCID: PMC11052255
- DOI: 10.3390/microorganisms12040695
Genomic and Phenotypic Analysis of Salmonella enterica Bacteriophages Identifies Two Novel Phage Species
Abstract
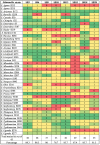
Bacteriophages (phages) are potential alternatives to chemical antimicrobials against pathogens of public health significance. Understanding the diversity and host specificity of phages is important for developing effective phage biocontrol approaches. Here, we assessed the host range, morphology, and genetic diversity of eight Salmonella enterica phages isolated from a wastewater treatment plant. The host range analysis revealed that six out of eight phages lysed more than 81% of the 43 Salmonella enterica isolates tested. The genomic sequences of all phages were determined. Whole-genome sequencing (WGS) data revealed that phage genome sizes ranged from 41 to 114 kb, with GC contents between 39.9 and 50.0%. Two of the phages SB13 and SB28 represent new species, Epseptimavirus SB13 and genera Macdonaldcampvirus, respectively, as designated by the International Committee for the Taxonomy of Viruses (ICTV) using genome-based taxonomic classification. One phage (SB18) belonged to the Myoviridae morphotype while the remaining phages belonged to the Siphoviridae morphotype. The gene content analyses showed that none of the phages possessed virulence, toxin, antibiotic resistance, type I-VI toxin-antitoxin modules, or lysogeny genes. Three (SB3, SB15, and SB18) out of the eight phages possessed tailspike proteins. Whole-genome-based phylogeny of the eight phages with their 113 homologs revealed three clusters A, B, and C and seven subclusters (A1, A2, A3, B1, B2, C1, and C2). While cluster C1 phages were predominantly isolated from animal sources, cluster B contained phages from both wastewater and animal sources. The broad host range of these phages highlights their potential use for controlling the presence of S. enterica in foods.
Keywords: Salmonella; bacteriophage; comparative phylogenomics; diversity; food safety; host specificity.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures




Similar articles
-
Diversity and Host Specificity Revealed by Biological Characterization and Whole Genome Sequencing of Bacteriophages Infecting Salmonella enterica.Viruses. 2019 Sep 14;11(9):854. doi: 10.3390/v11090854. Viruses. 2019. PMID: 31540091 Free PMC article.
-
Characterization of a Diverse Collection of Salmonella Phages Isolated from Tennessee Wastewater.Phage (New Rochelle). 2023 Jun 1;4(2):90-98. doi: 10.1089/phage.2023.0004. Epub 2023 Jun 19. Phage (New Rochelle). 2023. PMID: 37350991 Free PMC article.
-
Diversity of Salmonella enterica phages isolated from chicken farms in Kenya.Microbiol Spectr. 2024 Jan 11;12(1):e0272923. doi: 10.1128/spectrum.02729-23. Epub 2023 Dec 11. Microbiol Spectr. 2024. PMID: 38078723 Free PMC article.
-
Limitation of the Lytic Effect of Bacteriophages on Salmonella and Other Enteric Bacterial Pathogens and Approaches to Overcome.Int J Microbiol. 2025 May 15;2025:5936070. doi: 10.1155/ijm/5936070. eCollection 2025. Int J Microbiol. 2025. PMID: 40405891 Free PMC article. Review.
-
[Bacteriophages and bacteriocins of the genus Listeria].Zentralbl Bakteriol Mikrobiol Hyg A. 1986 Feb;261(1):12-28. Zentralbl Bakteriol Mikrobiol Hyg A. 1986. PMID: 3085381 Review. French.
Cited by
-
TSPDB: a curated resource of tailspike proteins with potential applications in phage research.Front Big Data. 2024 Nov 27;7:1437580. doi: 10.3389/fdata.2024.1437580. eCollection 2024. Front Big Data. 2024. PMID: 39664372 Free PMC article. No abstract available.
-
Recent Progress in Terrestrial Biota Derived Antibacterial Agents for Medical Applications.Molecules. 2024 Oct 15;29(20):4889. doi: 10.3390/molecules29204889. Molecules. 2024. PMID: 39459256 Free PMC article. Review.
-
Characterization and therapeutic evaluation of the lytic bacteriophage ENP2309 against vancomycin-resistant Enterococcus faecalis infections in a mice model.Virol J. 2025 Aug 28;22(1):292. doi: 10.1186/s12985-025-02917-1. Virol J. 2025. PMID: 40877963 Free PMC article.
References
-
- Thomas M.K., Murray R., Flockhart L., Pintar K., Fazil A., Nesbitt A., Marshall B., Tataryn J., Pollari F. Estimates of foodborne illness–related hospitalizations and deaths in Canada for 30 specified pathogens and unspecified agents. Foodborne Pathog. Dis. 2015;12:820–827. doi: 10.1089/fpd.2015.1966. - DOI - PMC - PubMed
-
- Majowicz S.E., Musto J., Scallan E., Angulo F.J., Kirk M., O’Brien S.J., Jones T.F., Fazil A., Hoekstra R.M., International Collaboration on Enteric Disease “Burden of Illness” Studies The global burden of nontyphoidal Salmonella gastroenteritis. Clin. Infect. Dis. 2010;50:882–889. doi: 10.1086/650733. - DOI - PubMed
-
- Grimont P.A.D., Weill F.-X. Antigenic Formulae of the Salmonella Serovars. 9th Edition. 2007. [(accessed on 13 February 2023)]. Available online: http://www.scacm.org/free/Antigenic%20Formulae%20of%20the%20Salmonella%2....
-
- PHAC Public Health Notice by Public Health Agency of Canada: Outbreak of Salmonella Infections Linked to Peaches Imported from the United States. 2020. [(accessed on 30 December 2023)]. Available online: http://tinyurl.com/44wz9p9v.
-
- CFIA Food Safety Investigation by Canadian Food Inspection Agency: Outbreak of Salmonella Infections Linked to Red Onions Imported from the United States. 2020. [(accessed on 30 December 2023)]. Available online: http://tinyurl.com/bddaefyn.
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

