Differential Selection for Translation Efficiency Shapes Translation Machineries in Bacterial Species
- PMID: 38674712
- PMCID: PMC11052298
- DOI: 10.3390/microorganisms12040768
Differential Selection for Translation Efficiency Shapes Translation Machineries in Bacterial Species
Abstract
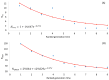
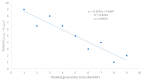
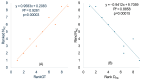
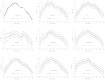
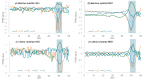
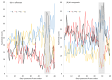
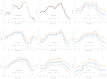
Different bacterial species have dramatically different generation times, from 20-30 min in Escherichia coli to about two weeks in Mycobacterium leprae. The translation machinery in a cell needs to synthesize all proteins for a new cell in each generation. The three subprocesses of translation, i.e., initiation, elongation, and termination, are expected to be under stronger selection pressure to optimize in short-generation bacteria (SGB) such as Vibrio natriegens than in the long-generation Mycobacterium leprae. The initiation efficiency depends on the start codon decoded by the initiation tRNA, the optimal Shine-Dalgarno (SD) decoded by the anti-SD (aSD) sequence on small subunit rRNA, and the secondary structure that may embed the initiation signals and prevent them from being decoded. The elongation efficiency depends on the tRNA pool and codon usage. The termination efficiency in bacteria depends mainly on the nature of the stop codon and the nucleotide immediately downstream of the stop codon. By contrasting SGB with long-generation bacteria (LGB), we predict (1) SGB to have more ribosome RNA operons to produce ribosomes, and more tRNA genes for carrying amino acids to ribosomes, (2) SGB to have a higher percentage of genes using AUG as the start codon and UAA as the stop codon than LGB, (3) SGB to exhibit better codon and anticodon adaptation than LGB, and (4) SGB to have a weaker secondary structure near the translation initiation signals than LGB. These differences between SGB and LGB should be more pronounced in highly expressed genes than the rest of the genes. We present empirical evidence in support of these predictions.
Keywords: Mycobacterium leprae; Mycobacterium tuberculosis; RNA secondary structure; rrn operons; tRNA; translation efficiency; translation elongation; translation initiation; translation termination.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures
Similar articles
-
Escherichia coli and Staphylococcus phages: effect of translation initiation efficiency on differential codon adaptation mediated by virulent and temperate lifestyles.J Gen Virol. 2015 May;96(Pt 5):1169-1179. doi: 10.1099/vir.0.000050. Epub 2015 Jan 22. J Gen Virol. 2015. PMID: 25614589 Free PMC article.
-
Initiator AUGs Are Discriminated from Elongator AUGs Predominantly through mRNA Accessibility in C. crescentus.J Bacteriol. 2023 May 25;205(5):e0042022. doi: 10.1128/jb.00420-22. Epub 2023 Apr 24. J Bacteriol. 2023. PMID: 37092987 Free PMC article.
-
Local absence of secondary structure permits translation of mRNAs that lack ribosome-binding sites.PLoS Genet. 2011 Jun;7(6):e1002155. doi: 10.1371/journal.pgen.1002155. Epub 2011 Jun 23. PLoS Genet. 2011. PMID: 21731509 Free PMC article.
-
The diversity of Shine-Dalgarno sequences sheds light on the evolution of translation initiation.RNA Biol. 2021 Nov;18(11):1489-1500. doi: 10.1080/15476286.2020.1861406. Epub 2020 Dec 21. RNA Biol. 2021. PMID: 33349119 Free PMC article. Review.
-
Start Codon Recognition in Eukaryotic and Archaeal Translation Initiation: A Common Structural Core.Int J Mol Sci. 2019 Feb 21;20(4):939. doi: 10.3390/ijms20040939. Int J Mol Sci. 2019. PMID: 30795538 Free PMC article. Review.
References
-
- Jacob F. Génétique Cellulaire: Leçon Inaugurale Prononcée le Vendredi 7 mai 1965. Collège de France; Paris, France: 2013.
Grants and funding
LinkOut - more resources
Full Text Sources

