Expanding the CRISPR Toolbox for Engineering Lycopene Biosynthesis in Corynebacterium glutamicum
- PMID: 38674747
- PMCID: PMC11052027
- DOI: 10.3390/microorganisms12040803
Expanding the CRISPR Toolbox for Engineering Lycopene Biosynthesis in Corynebacterium glutamicum
Abstract
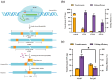
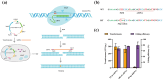
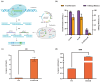
Lycopene represents one of the central compounds in the carotenoid pathway and it exhibits a potent antioxidant ability with wide potential applications in medicine, food, and cosmetics. The microbial production of lycopene has received increasing concern in recent years. Corynebacterium glutamicum (C. glutamicum) is considered to be a safe and beneficial industrial production platform, naturally endowed with the ability to produce lycopene. However, the scarcity of efficient genetic tools and the challenge of identifying crucial metabolic genes impede further research on C. glutamicum for achieving high-yield lycopene production. To address these challenges, a novel genetic editing toolkit, CRISPR/MAD7 system, was established and developed. By optimizing the promoter, ORI and PAM sequences, the CRISPR/MAD7 system facilitated highly efficient gene deletion and exhibited a broad spectrum of PAM sites. Notably, 25 kb of DNA from the genome was successfully deleted. In addition, the CRISPR/MAD7 system was effectively utilized in the metabolic engineering of C. glutamicum, allowing for the simultaneous knockout of crtEb and crtR genes in one step to enhance the accumulation of lycopene by blocking the branching pathway. Through screening crucial genes such as crtE, crtB, crtI, idsA, idi, and cg0722, an optimal carotenogenic gene combination was obtained. Particularly, cg0722, a membrane protein gene, was found to play a vital role in lycopene production. Therefore, the CBIEbR strain was obtained by overexpressing cg0722, crtB, and crtI while strategically blocking the by-products of the lycopene pathway. As a result, the final engineered strain produced lycopene at 405.02 mg/L (9.52 mg/g dry cell weight, DCW) in fed-batch fermentation, representing the highest reported lycopene yield in C. glutamicum to date. In this study, a powerful and precise genetic tool was used to engineer C. glutamicum for lycopene production. Through the modifications between the host cell and the carotenogenic pathway, the lycopene yield was stepwise improved by 102-fold as compared to the starting strain. This study highlights the usefulness of the CRISPR/MAD7 toolbox, demonstrating its practical applications in the metabolic engineering of industrially robust C. glutamicum.
Keywords: CRISPR/MAD7; Corynebacterium glutamicum; lycopene production; metabolic engineering.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures



Similar articles
-
Carotenoid biosynthesis and overproduction in Corynebacterium glutamicum.BMC Microbiol. 2012 Sep 10;12:198. doi: 10.1186/1471-2180-12-198. BMC Microbiol. 2012. PMID: 22963379 Free PMC article.
-
Lycopene overproduction in Saccharomyces cerevisiae through combining pathway engineering with host engineering.Microb Cell Fact. 2016 Jun 21;15(1):113. doi: 10.1186/s12934-016-0509-4. Microb Cell Fact. 2016. PMID: 27329233 Free PMC article.
-
Production of lycopene by metabolically engineered Pichia pastoris.Biosci Biotechnol Biochem. 2020 Mar;84(3):463-470. doi: 10.1080/09168451.2019.1693250. Epub 2019 Nov 22. Biosci Biotechnol Biochem. 2020. PMID: 31752618
-
[Recent advances in developing enabling technologies for Corynebacterium glutamicum metabolic engineering].Sheng Wu Gong Cheng Xue Bao. 2021 May 25;37(5):1603-1618. doi: 10.13345/j.cjb.200649. Sheng Wu Gong Cheng Xue Bao. 2021. PMID: 34085445 Review. Chinese.
-
Metabolic engineering of Corynebacterium glutamicum for producing branched chain amino acids.Microb Cell Fact. 2021 Dec 24;20(1):230. doi: 10.1186/s12934-021-01721-0. Microb Cell Fact. 2021. PMID: 34952576 Free PMC article. Review.
Cited by
-
Tailoring Corynebacterium glutamicum for Sustainable Biomanufacturing: From Traditional to Cutting-Edge Technologies.Mol Biotechnol. 2025 Jun 10. doi: 10.1007/s12033-025-01447-z. Online ahead of print. Mol Biotechnol. 2025. PMID: 40493161 Review.
References
-
- Wang Y.-H., Zhang R.-R., Yin Y., Tan G.-F., Wang G.-L., Liu H., Zhuang J., Zhang J., Zhuang F.-Y., Xiong A.-S. Advances in engineering the production of the natural red pigment lycopene: A systematic review from a biotechnology perspective. J. Adv. Res. 2023;46:31–47. doi: 10.1016/j.jare.2022.06.010. - DOI - PMC - PubMed
-
- Dupont S., Fleurat-Lessard P., Cruz R.G., Lafarge C., Grangeteau C., Yahou F., Gerbeau-Pissot P., Júnior O.A., Gervais P., Simon-Plas F., et al. Antioxidant properties of ergosterol and its role in yeast resistance to oxidation. Antioxidants. 2021;10:1024. doi: 10.3390/antiox10071024. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

