Evaluation of Probiotic Properties and Safety of Lactobacillus helveticus LH10 Derived from Vinegar through Comprehensive Analysis of Genotype and Phenotype
- PMID: 38674775
- PMCID: PMC11052092
- DOI: 10.3390/microorganisms12040831
Evaluation of Probiotic Properties and Safety of Lactobacillus helveticus LH10 Derived from Vinegar through Comprehensive Analysis of Genotype and Phenotype
Abstract
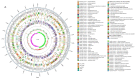
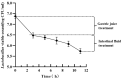
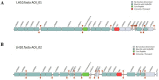
The probiotic potential of Lactobacillus helveticus LH10, derived from vinegar Pei, a brewing mixture, was assessed through genotype and phenotype analyses. The assembled genome was comprised of 1,810,276 bp and predicted a total of 2044 coding sequences (CDSs). Based on the whole genome sequence analysis, two bacteriocin gene clusters were identified, while no pathogenic genes were detected. In in vitro experiments, L. helveticus LH10 exhibited excellent tolerance to simulated gastrointestinal fluid, a positive hydrophobic interaction with xylene, and good auto-aggregation properties. Additionally, this strain demonstrated varying degrees of resistance to five antibiotics, strong antagonistic activity against four tested pathogens, and no hemolytic activity. Therefore, L. helveticus LH10 holds great promise as a potential probiotic candidate deserving further investigation for its beneficial effects on human health.
Keywords: Lactobacillus helveticus; cereal vinegar; phenotypic analysis; safety and probiotic properties; whole-genome sequencing.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures






References
-
- Pimentel T.C., Cruz A.G., Pereira E., Almeida Da Costa W.K., Da Silva Rocha R., Targino De Souza Pedrosa G., Rocha C.D.S., Alves J.M., Alvarenga V.O., Sant’Ana A.S., et al. Postbiotics: An overview of concepts, inactivation technologies, health effects, and driver trends. Trends Food Sci. Technol. 2023;138:199–214. doi: 10.1016/j.tifs.2023.06.009. - DOI
-
- Takeda T., Asaoka D., Nojiri S., Yanagisawa N., Nishizaki Y., Osada T., Koido S., Nagahara A., Katsumata N., Odamaki T., et al. Usefulness of Bifidobacterium longum BB536 in Elderly Individuals With Chronic Constipation: A Randomized Controlled Trial. Off. J. Am. Coll. Gastroenterol. ACG. 2023;118:561. doi: 10.14309/ajg.0000000000002028. - DOI - PMC - PubMed
-
- Forsgård R.A., Rode J., Lobenius-Palmér K., Kamm A., Patil S., Tacken M.G.J., Lentjes M.A.H., Axelsson J., Grompone G., Montgomery S., et al. Limosilactobacillus reuteri DSM 17938 Supplementation and SARS-CoV-2 Specific Antibody Response in Healthy Adults: A Randomized, Triple-Blinded, Placebo-Controlled Trial. Gut Microbes. 2023;15:2229938. doi: 10.1080/19490976.2023.2229938. - DOI - PMC - PubMed
-
- Fan M., Guo T., Li W., Chen J., Li F., Wang C., Shi Y., Li D.X., Zhang S. Isolation and Identification of Novel Casein-Derived Bioactive Peptides and Potential Functions in Fermented Casein with Lactobacillus Helveticus. Food Sci. Hum. Wellness. 2019;8:156–176. doi: 10.1016/j.fshw.2019.03.010. - DOI
Grants and funding
LinkOut - more resources
Full Text Sources

