The Development of Aptamer-Based Gold Nanoparticle Lateral Flow Test Strips for the Detection of SARS-CoV-2 S Proteins on the Surface of Cold-Chain Food Packaging
- PMID: 38675595
- PMCID: PMC11052266
- DOI: 10.3390/molecules29081776
The Development of Aptamer-Based Gold Nanoparticle Lateral Flow Test Strips for the Detection of SARS-CoV-2 S Proteins on the Surface of Cold-Chain Food Packaging
Abstract
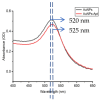
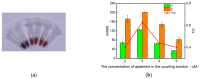
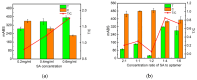
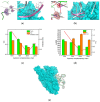
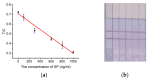
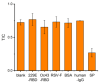
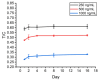
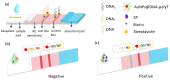
The COVID-19 pandemic over recent years has shown a great need for the rapid, low-cost, and on-site detection of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). In this study, an aptamer-based colloidal gold nanoparticle lateral flow test strip was well developed to realize the visual detection of wild-type SARS-CoV-2 spike proteins (SPs) and multiple variants. Under the optimal reaction conditions, a low detection limit of SARS-CoV-2 S proteins of 0.68 nM was acquired, and the actual detection recovery was 83.3% to 108.8% for real-world samples. This suggests a potential tool for the prompt detection of SARS-CoV-2 with good sensitivity and accuracy, and a new method for the development of alternative antibody test strips for the detection of other viral targets.
Keywords: AuNPs; COVID-19; S protein; SARS-CoV-2; aptamer; lateral flow test strips.
Conflict of interest statement
The authors declare no conflicts of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Figures



References
-
- World Health Organization WHO Coronavirus (COVID-19) Dashboard. [(accessed on 17 January 2024)];2024 Available online: https://covid19.who.int/
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

