Modeling Studies of the Mechanism of Context-Dependent Bidirectional Movements of Kinesin-14 Motors
- PMID: 38675612
- PMCID: PMC11055046
- DOI: 10.3390/molecules29081792
Modeling Studies of the Mechanism of Context-Dependent Bidirectional Movements of Kinesin-14 Motors
Abstract
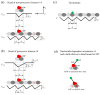
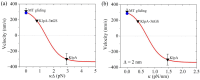
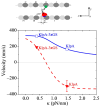
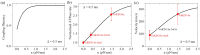
Kinesin-14s, a subfamily of the large superfamily of kinesin motor proteins, function mainly in spindle assembly and maintenance during mitosis and meiosis. KlpA from Aspergillus nidulans and GiKIN14a from Giardia intestinalis are two types of kinesin-14s. Available experimental results puzzlingly showed that while KlpA moves preferentially toward the minus end in microtubule-gliding setups and inside parallel microtubule overlaps, it moves preferentially toward the plus end on single microtubules. More puzzlingly, the insertion of an extra polypeptide linker in the central region of the neck stalk switches the motility direction of KlpA on single microtubules to the minus end. Prior experimental results showed that GiKIN14a moves preferentially toward the minus end on single microtubules in either tailless or full-length forms. The tail not only greatly enhances the processivity but also accelerates the ATPase rate and velocity of GiKIN14a. The insertion of an extra polypeptide linker in the central region of the neck stalk reduces the ATPase rate of GiKIN14a. However, the underlying mechanism of these puzzling dynamical features for KlpA and GiKIN14a is unclear. Here, to understand this mechanism, the dynamics of KlpA and GiKIN14a were studied theoretically on the basis of the proposed model, incorporating potential changes between the kinesin head and microtubule, as well as the potential between the tail and microtubule. The theoretical results quantitatively explain the available experimental results and provide predicted results. It was found that the elasticity of the neck stalk determines the directionality of KlpA on single microtubules and affects the ATPase rate and velocity of GiKIN14a on single microtubules.
Keywords: chemo–mechanical coupling mechanism; kinesin; molecular motor; movement direction.
Conflict of interest statement
The author declares no conflicts of interest.
Figures



Similar articles
-
The Tail of Kinesin-14a in Giardia Is a Dual Regulator of Motility.Curr Biol. 2020 Sep 21;30(18):3664-3671.e4. doi: 10.1016/j.cub.2020.06.090. Epub 2020 Jul 30. Curr Biol. 2020. PMID: 32735815 Free PMC article.
-
The mitotic kinesin-14 KlpA contains a context-dependent directionality switch.Nat Commun. 2017 Jan 4;8:13999. doi: 10.1038/ncomms13999. Nat Commun. 2017. PMID: 28051135 Free PMC article.
-
The Central Stalk Determines the Motility of Mitotic Kinesin-14 Homodimers.Curr Biol. 2018 Jul 23;28(14):2302-2308.e3. doi: 10.1016/j.cub.2018.05.026. Epub 2018 Jul 12. Curr Biol. 2018. PMID: 30017487
-
A model of microtubule depolymerization by kinesin-8 motor proteins.Adv Protein Chem Struct Biol. 2024;141:87-122. doi: 10.1016/bs.apcsb.2023.12.002. Epub 2023 Dec 28. Adv Protein Chem Struct Biol. 2024. PMID: 38960488 Review.
-
Bidirectional motility of kinesin-5 motor proteins: structural determinants, cumulative functions and physiological roles.Cell Mol Life Sci. 2018 May;75(10):1757-1771. doi: 10.1007/s00018-018-2754-7. Epub 2018 Feb 3. Cell Mol Life Sci. 2018. PMID: 29397398 Free PMC article. Review.
References
MeSH terms
LinkOut - more resources
Full Text Sources

