Natural Killer Cells Do Not Attenuate a Mouse-Adapted SARS-CoV-2-Induced Disease in Rag2-/- Mice
- PMID: 38675952
- PMCID: PMC11054502
- DOI: 10.3390/v16040611
Natural Killer Cells Do Not Attenuate a Mouse-Adapted SARS-CoV-2-Induced Disease in Rag2-/- Mice
Abstract
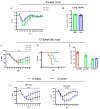
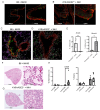
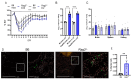
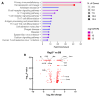
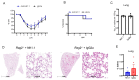
This study investigates the roles of T, B, and Natural Killer (NK) cells in the pathogenesis of severe COVID-19, utilizing mouse-adapted SARS-CoV-2-MA30 (MA30). To evaluate this MA30 mouse model, we characterized MA30-infected C57BL/6 mice (B6) and compared them with SARS-CoV-2-WA1 (an original SARS-CoV-2 strain) infected K18-human ACE2 (K18-hACE2) mice. We found that the infected B6 mice developed severe peribronchial inflammation and rapid severe pulmonary edema, but less lung interstitial inflammation than the infected K18-hACE2 mice. These pathological findings recapitulate some pathological changes seen in severe COVID-19 patients. Using this MA30-infected mouse model, we further demonstrate that T and/or B cells are essential in mounting an effective immune response against SARS-CoV-2. This was evident as Rag2-/- showed heightened vulnerability to infection and inhibited viral clearance. Conversely, the depletion of NK cells did not significantly alter the disease course in Rag2-/- mice, underscoring the minimal role of NK cells in the acute phase of MA30-induced disease. Together, our results indicate that T and/or B cells, but not NK cells, mitigate MA30-induced disease in mice and the infected mouse model can be used for dissecting the pathogenesis and immunology of severe COVID-19.
Keywords: B cells; COVID-19; MA30; NK cells; SARS-CoV-2; T cells.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures
References
-
- Fernandes Q., Inchakalody V.P., Merhi M., Mestiri S., Taib N., Moustafa Abo El-Ella D., Bedhiafi T., Raza A., Al-Zaidan L., Mohsen M.O., et al. Emerging COVID-19 variants and their impact on SARS-CoV-2 diagnosis, therapeutics and vaccines. Ann. Med. 2022;54:524–540. doi: 10.1080/07853890.2022.2031274. - DOI - PMC - PubMed
-
- Malahe S.R.K., Hoek R.A.S., Dalm V., Broers A.E.C., den Hoed C.M., Manintveld O.C., Baan C.C., van Deuzen C.M., Papageorgiou G., Bax H.I., et al. Clinical Characteristics and Outcomes of Immunocompromised Patients with Coronavirus Disease 2019 Caused by the Omicron Variant: A Prospective, Observational Study. Clin. Infect. Dis. 2023;76:e172–e178. doi: 10.1093/cid/ciac571. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

