The Dual-Targeted Fusion Inhibitor Clofazimine Binds to the S2 Segment of the SARS-CoV-2 Spike Protein
- PMID: 38675980
- PMCID: PMC11054727
- DOI: 10.3390/v16040640
The Dual-Targeted Fusion Inhibitor Clofazimine Binds to the S2 Segment of the SARS-CoV-2 Spike Protein
Abstract
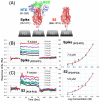
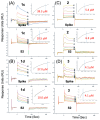
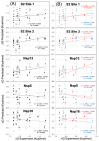
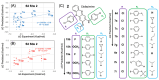
Clofazimine and Arbidol have both been reported to be effective in vitro SARS-CoV-2 fusion inhibitors. Both are promising drugs that have been repurposed for the treatment of COVID-19 and have been used in several previous and ongoing clinical trials. Small-molecule bindings to expressed constructs of the trimeric S2 segment of Spike and the full-length SARS-CoV-2 Spike protein were measured using a Surface Plasmon Resonance (SPR) binding assay. We demonstrate that Clofazimine, Toremifene, Arbidol and its derivatives bind to the S2 segment of the Spike protein. Clofazimine provided the most reliable and highest-quality SPR data for binding with S2 over the conditions explored. A molecular docking approach was used to identify the most favorable binding sites on the S2 segment in the prefusion conformation, highlighting two possible small-molecule binding sites for fusion inhibitors. Results related to molecular docking and modeling of the structure-activity relationship (SAR) of a newly reported series of Clofazimine derivatives support the proposed Clofazimine binding site on the S2 segment. When the proposed Clofazimine binding site is superimposed with other experimentally determined coronavirus structures in structure-sequence alignments, the changes in sequence and structure may rationalize the broad-spectrum antiviral activity of Clofazimine in closely related coronaviruses such as SARS-CoV, MERS, hCoV-229E, and hCoV-OC43.
Keywords: Arbidol; CHARMM; Clofazimine; Nsp13 helicase; S2 segment; S2 subunit; SARS-CoV-2; Spike-dependent; Toremifene; fusion inhibitor; molecular docking; surface plasmon resonance.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures






References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

