RecGraph: recombination-aware alignment of sequences to variation graphs
- PMID: 38676570
- PMCID: PMC11256948
- DOI: 10.1093/bioinformatics/btae292
RecGraph: recombination-aware alignment of sequences to variation graphs
Abstract
Motivation: Bacterial genomes present more variability than human genomes, which requires important adjustments in computational tools that are developed for human data. In particular, bacteria exhibit a mosaic structure due to homologous recombinations, but this fact is not sufficiently captured by standard read mappers that align against linear reference genomes. The recent introduction of pangenomics provides some insights in that context, as a pangenome graph can represent the variability within a species. However, the concept of sequence-to-graph alignment that captures the presence of recombinations has not been previously investigated.
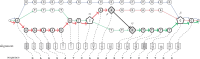
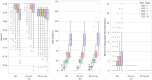
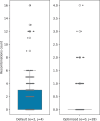
Results: In this paper, we present the extension of the notion of sequence-to-graph alignment to a variation graph that incorporates a recombination, so that the latter are explicitly represented and evaluated in an alignment. Moreover, we present a dynamic programming approach for the special case where there is at most a recombination-we implement this case as RecGraph. From a modelling point of view, a recombination corresponds to identifying a new path of the variation graph, where the new arc is composed of two halves, each extracted from an original path, possibly joined by a new arc. Our experiments show that RecGraph accurately aligns simulated recombinant bacterial sequences that have at most a recombination, providing evidence for the presence of recombination events.
Availability and implementation: Our implementation is open source and available at https://github.com/AlgoLab/RecGraph.
© The Author(s) 2024. Published by Oxford University Press.
Conflict of interest statement
None declared.
Figures

References
-
- Amir A, Lewenstein M, Lewenstein N. Pattern matching in hypertext. J Algorithms 2000;35:82–99.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

