Sea Anemone Membrane Attack Complex/Perforin Superfamily Demonstrates an Evolutionary Transitional State between Venomous and Developmental Functions
- PMID: 38676945
- PMCID: PMC11090067
- DOI: 10.1093/molbev/msae082
Sea Anemone Membrane Attack Complex/Perforin Superfamily Demonstrates an Evolutionary Transitional State between Venomous and Developmental Functions
Abstract
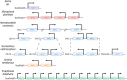
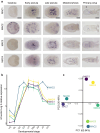
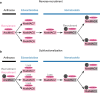
Gene duplication is a major force driving evolutionary innovation. A classic example is generating new animal toxins via duplication of physiological protein-encoding genes and recruitment into venom. While this process drives the innovation of many animal venoms, reverse recruitment of toxins into nonvenomous cells remains unresolved. Using comparative genomics, we find members of the Membrane Attack Complex and Perforin Family (MAC) have been recruited into venom-injecting cells (cnidocytes), in soft and stony corals and sea anemones, suggesting that the ancestral MAC was a cnidocyte expressed toxin. Further investigation into the model sea anemone Nematostella vectensis reveals that three members have undergone Nematostella-specific duplications leading to their reverse recruitment into endomesodermal cells. Furthermore, simultaneous knockdown of all three endomesodermally expressed MACs leads to mis-development, supporting that these paralogs have nonvenomous function. By resolving the evolutionary history and function of MACs in Nematostella, we provide the first proof for reverse recruitment from venom to organismal development.
Keywords: Cnidaria; gene duplication; reverse recruitment; subfunctionalization; toxin; venom.
© The Author(s) 2024. Published by Oxford University Press on behalf of Society for Molecular Biology and Evolution.
Figures


Similar articles
-
Toxin-like neuropeptides in the sea anemone Nematostella unravel recruitment from the nervous system to venom.Proc Natl Acad Sci U S A. 2020 Nov 3;117(44):27481-27492. doi: 10.1073/pnas.2011120117. Epub 2020 Oct 15. Proc Natl Acad Sci U S A. 2020. PMID: 33060291 Free PMC article.
-
The Birth and Death of Toxins with Distinct Functions: A Case Study in the Sea Anemone Nematostella.Mol Biol Evol. 2019 Sep 1;36(9):2001-2012. doi: 10.1093/molbev/msz132. Mol Biol Evol. 2019. PMID: 31134275
-
Evolution of an ancient venom: recognition of a novel family of cnidarian toxins and the common evolutionary origin of sodium and potassium neurotoxins in sea anemone.Mol Biol Evol. 2015 Jun;32(6):1598-610. doi: 10.1093/molbev/msv050. Epub 2015 Mar 9. Mol Biol Evol. 2015. PMID: 25757852
-
Never, Ever Make an Enemy… Out of an Anemone: Transcriptomic Comparison of Clownfish Hosting Sea Anemone Venoms.Mar Drugs. 2022 Nov 23;20(12):730. doi: 10.3390/md20120730. Mar Drugs. 2022. PMID: 36547877 Free PMC article. Review.
-
Towards the Exploration and Evolution of Insulin-like Venoms in Actiniaria (Sea anemones).Mar Drugs. 2024 Mar 20;22(3):136. doi: 10.3390/md22030136. Mar Drugs. 2024. PMID: 38535477 Free PMC article. Review.
Cited by
-
The Hydractinia cell atlas reveals cellular and molecular principles of cnidarian coloniality.Nat Commun. 2025 Mar 3;16(1):2121. doi: 10.1038/s41467-025-57168-z. Nat Commun. 2025. PMID: 40032860 Free PMC article.
-
A comparative analysis of toxin gene families across diverse sea anemone species.Toxicon X. 2025 Mar 7;26:100217. doi: 10.1016/j.toxcx.2025.100217. eCollection 2025 Jun. Toxicon X. 2025. PMID: 40162058 Free PMC article.
References
-
- Al-Shaer L, Havrilak J, Layden MJ. Nematostella vectensis as a model system. In: Handbook of marine model organisms in experimental biology. 1st ed. Boca Raton: CRC Press; 2021. p. 107–128
-
- Anderluh G, Kisovec M, Kraševec N, Gilbert RJC. Distribution of MAC/CDC proteins. In: Anderluh G, Gilbert R, editors. MAC/CDC proteins—agents of defence, attack and invasion. Subcellular biochemistry. Dordrecht: Springer Netherlands; 2014. p. 7–30.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

