Endogenous retroelement expression in the gut microenvironment of people living with HIV-1
- PMID: 38677181
- PMCID: PMC11061259
- DOI: 10.1016/j.ebiom.2024.105133
Endogenous retroelement expression in the gut microenvironment of people living with HIV-1
Abstract
Background: Endogenous retroelements (EREs), including human endogenous retroviruses (HERVs) and long interspersed nuclear elements (LINEs), comprise almost half of the human genome. Our previous studies of the interferome in the gut suggest potential mechanisms regarding how IFNb may drive HIV-1 gut pathogenesis. As ERE activity is suggested to partake in type 1 immune responses and is incredibly sensitive to viral infections, we sought to elucidate underlying interactions between ERE expression and gut dynamics in people living with HIV-1 (PLWH).
Methods: ERE expression profiles from bulk RNA sequencing of colon biopsies and PBMC were compared between a cohort of PLWH not on antiretroviral therapy (ART) and uninfected controls.
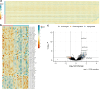
Findings: 59 EREs were differentially expressed in the colon of PLWH when compared to uninfected controls (padj <0.05 and FC ≤ -1 or ≥ 1) [Wald's Test]. Of these 59, 12 EREs were downregulated in PLWH and 47 were upregulated. Colon expression of the ERE loci LTR19_12p13.31 and L1FLnI_1q23.1s showed significant correlations with certain gut immune cell subset frequencies in the colon. Furthermore L1FLnI_1q23.1s showed a significant upregulation in peripheral blood mononuclear cells (PBMCs) of PLWH when compared to uninfected controls suggesting a common mechanism of differential ERE expression in the colon and PBMC.
Interpretation: ERE activity has been largely understudied in genomic characterizations of human pathologies. We show that the activity of certain EREs in the colon of PLWH is deregulated, supporting our hypotheses that their underlying activity could function as (bio)markers and potential mediators of pathogenesis in HIV-1 reservoirs.
Funding: US NIH grants NCI CA260691 (DFN) and NIAID UM1AI164559 (DFN).
Keywords: Endogenous retroelement (ERE); Human endogenous retrovirus (HERV); Human immunodeficiency virus type I (HIV-I); Interferons; Long interspersed nuclear element (LINE).
Copyright © 2024 The Author(s). Published by Elsevier B.V. All rights reserved.
Conflict of interest statement
Declaration of interests All authors have no potential conflicts of interests to disclose.
Figures


References
-
- Justiz Vaillant A.A., Gulick P.G. StatPearls. StatPearls Publishing; Treasure Island (FL): 2023. HIV disease current practice.
-
- Deeks S.G., Overbaugh J., Phillips A., Buchbinder S. HIV infection. Nat. Rev Dis Primer. 2015;1:1–22. - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

