The hinge-engineered IgG1-IgG3 hybrid subclass IgGh47 potently enhances Fc-mediated function of anti-streptococcal and SARS-CoV-2 antibodies
- PMID: 38678029
- PMCID: PMC11055898
- DOI: 10.1038/s41467-024-47928-8
The hinge-engineered IgG1-IgG3 hybrid subclass IgGh47 potently enhances Fc-mediated function of anti-streptococcal and SARS-CoV-2 antibodies
Abstract
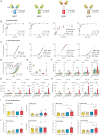
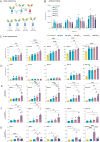
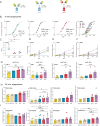
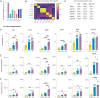
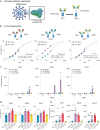
Streptococcus pyogenes can cause invasive disease with high mortality despite adequate antibiotic treatments. To address this unmet need, we have previously generated an opsonic IgG1 monoclonal antibody, Ab25, targeting the bacterial M protein. Here, we engineer the IgG2-4 subclasses of Ab25. Despite having reduced binding, the IgG3 version promotes stronger phagocytosis of bacteria. Using atomic simulations, we show that IgG3's Fc tail has extensive movement in 3D space due to its extended hinge region, possibly facilitating interactions with immune cells. We replaced the hinge of IgG1 with four different IgG3-hinge segment subclasses, IgGhxx. Hinge-engineering does not diminish binding as with IgG3 but enhances opsonic function, where a 47 amino acid hinge is comparable to IgG3 in function. IgGh47 shows improved protection against S. pyogenes in a systemic infection mouse model, suggesting that IgGh47 has promise as a preclinical therapeutic candidate. Importantly, the enhanced opsonic function of IgGh47 is generalizable to diverse S. pyogenes strains from clinical isolates. We generated IgGh47 versions of anti-SARS-CoV-2 mAbs to broaden the biological applicability, and these also exhibit strongly enhanced opsonic function compared to the IgG1 subclass. The improved function of the IgGh47 subclass in two distant biological systems provides new insights into antibody function.
© 2024. The Author(s).
Conflict of interest statement
AI, HK, WB, LH, OS, JM, LM, and PN have filed patents related to the mAbs described in this manuscript. The remaining authors declare no competing interests.
Figures



Similar articles
-
Subclass-switched anti-spike IgG3 oligoclonal cocktails strongly enhance Fc-mediated opsonization.Proc Natl Acad Sci U S A. 2023 Apr 11;120(15):e2217590120. doi: 10.1073/pnas.2217590120. Epub 2023 Apr 3. Proc Natl Acad Sci U S A. 2023. PMID: 37011197 Free PMC article.
-
IgG1 versus IgG3: influence of antibody-specificity and allotypic variance on virus neutralization efficacy.Front Immunol. 2024 Oct 24;15:1490515. doi: 10.3389/fimmu.2024.1490515. eCollection 2024. Front Immunol. 2024. PMID: 39512357 Free PMC article.
-
The Proteolytic Cleavage of Therapeutic Monoclonal Antibody Hinge Region: More Than a Matter of Subclass.Front Immunol. 2020 Feb 11;11:168. doi: 10.3389/fimmu.2020.00168. eCollection 2020. Front Immunol. 2020. PMID: 32117299 Free PMC article.
-
Human IgG1, IgG3, and IgG3 Hinge-Truncated Mutants Show Different Protection Capabilities against Meningococci Depending on the Target Antigen and Epitope Specificity.Clin Vaccine Immunol. 2016 Aug 5;23(8):698-706. doi: 10.1128/CVI.00193-16. Print 2016 Aug. Clin Vaccine Immunol. 2016. PMID: 27307451 Free PMC article.
-
Engineered IgG1-Fc--one fragment to bind them all.Immunol Rev. 2016 Mar;270(1):113-31. doi: 10.1111/imr.12385. Immunol Rev. 2016. PMID: 26864108 Free PMC article. Review.
Cited by
-
Dissecting the properties of circulating IgG against streptococcal pathogens through a combined systems antigenomics-serology workflow.Nat Commun. 2025 Feb 24;16(1):1942. doi: 10.1038/s41467-025-57170-5. Nat Commun. 2025. PMID: 39994218 Free PMC article.
-
Protective Non-neutralizing anti-N-terminal Domain mAb Maintains Fc-mediated Function against SARS-COV-2 Variants up to BA.2.86-JN.1 with Superfluous In Vivo Protection against JN.1 Due to Attenuated Virulence.J Immunol. 2024 Sep 1;213(5):678-689. doi: 10.4049/jimmunol.2300675. J Immunol. 2024. PMID: 39018495 Free PMC article.
-
Stellabody: A novel hexamer-promoting mutation for improved IgG potency.Immunol Rev. 2024 Nov;328(1):438-455. doi: 10.1111/imr.13400. Epub 2024 Oct 4. Immunol Rev. 2024. PMID: 39364646 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

