Lifecycle of a predatory bacterium vampirizing its prey through the cell envelope and S-layer
- PMID: 38678033
- PMCID: PMC11055950
- DOI: 10.1038/s41467-024-48042-5
Lifecycle of a predatory bacterium vampirizing its prey through the cell envelope and S-layer
Erratum in
-
Author Correction: Lifecycle of a predatory bacterium vampirizing its prey through the cell envelope and S-layer.Nat Commun. 2025 Apr 15;16(1):3567. doi: 10.1038/s41467-025-58758-7. Nat Commun. 2025. PMID: 40234419 Free PMC article. No abstract available.
Abstract
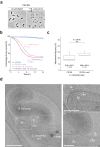
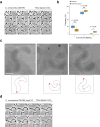
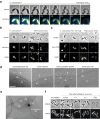
Predatory bacteria feed upon other bacteria in various environments. Bdellovibrio exovorus is an obligate epibiotic predator that attaches on the prey cell surface, where it grows and proliferates. Although the mechanisms allowing feeding through the prey cell envelope are unknown, it has been proposed that the prey's proteinaceous S-layer may act as a defensive structure against predation. Here, we use time-lapse and cryo-electron microscopy to image the lifecycle of B. exovorus feeding on Caulobacter crescentus. We show that B. exovorus proliferates by non-binary division, primarily generating three daughter cells. Moreover, the predator feeds on C. crescentus regardless of the presence of an S-layer, challenging its assumed protective role against predators. Finally, we show that apparently secure junctions are established between prey and predator outer membranes.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
- ERC Starting Grant PREDATOR #802331/EC | EU Framework Programme for Research and Innovation H2020 | H2020 Priority Excellent Science | H2020 European Research Council (H2020 Excellent Science - European Research Council)
- Post-doctoral fellowship/European Molecular Biology Organization (EMBO)
- Post-doctoral fellowship/European Molecular Biology Organization (EMBO)
- Post-doctoral fellowship/Fonds De La Recherche Scientifique - FNRS (Belgian National Fund for Scientific Research)
- Post-doctoral fellowship/EC | EU Framework Programme for Research and Innovation H2020 | H2020 Priority Excellent Science | H2020 Marie Skłodowska-Curie Actions (H2020 Excellent Science - Marie Skłodowska-Curie Actions)
LinkOut - more resources
Full Text Sources

