High-resolution genomics identifies pneumococcal diversity and persistence of vaccine types in children with community-acquired pneumonia in the UK and Ireland
- PMID: 38678217
- PMCID: PMC11055344
- DOI: 10.1186/s12866-024-03300-w
High-resolution genomics identifies pneumococcal diversity and persistence of vaccine types in children with community-acquired pneumonia in the UK and Ireland
Abstract
Background: Streptococcus pneumoniae is a global cause of community-acquired pneumonia (CAP) and invasive disease in children. The CAP-IT trial (grant No. 13/88/11; https://www.capitstudy.org.uk/ ) collected nasopharyngeal swabs from children discharged from hospitals with clinically diagnosed CAP, and found no differences in pneumococci susceptibility between higher and lower antibiotic doses and shorter and longer durations of oral amoxicillin treatment. Here, we studied in-depth the genomic epidemiology of pneumococcal (vaccine) serotypes and their antibiotic resistance profiles.
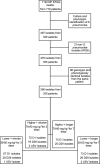
Methods: Three-hundred and ninety pneumococci cultured from 1132 nasopharyngeal swabs from 718 children were whole-genome sequenced (Illumina) and tested for susceptibility to penicillin and amoxicillin. Genome heterogeneity analysis was performed using long-read sequenced isolates (PacBio, n = 10) and publicly available sequences.
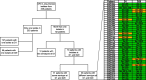
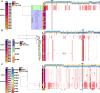
Results: Among 390 unique pneumococcal isolates, serotypes 15B/C, 11 A, 15 A and 23B1 were most prevalent (n = 145, 37.2%). PCV13 serotypes 3, 19A, and 19F were also identified (n = 25, 6.4%). STs associated with 19A and 19F demonstrated high genome variability, in contrast to serotype 3 (n = 13, 3.3%) that remained highly stable over a 20-year period. Non-susceptibility to penicillin (n = 61, 15.6%) and amoxicillin (n = 10, 2.6%) was low among the pneumococci analysed here and was independent of treatment dosage and duration. However, all 23B1 isolates (n = 27, 6.9%) were penicillin non-susceptible. This serotype was also identified in ST177, which is historically associated with the PCV13 serotype 19F and penicillin susceptibility, indicating a potential capsule-switch event.
Conclusions: Our data suggest that amoxicillin use does not drive pneumococcal serotype prevalence among children in the UK, and prompts consideration of PCVs with additional serotype coverage that are likely to further decrease CAP in this target population. Genotype 23B1 represents the convergence of a non-vaccine genotype with penicillin non-susceptibility and might provide a persistence strategy for ST types historically associated with vaccine serotypes. This highlights the need for continued genomic surveillance.
Keywords: Streptococcus pneumoniae; Beta-lactams; Emerging serotype; Non-vaccine types; Population genomics; Vaccine escape; Vaccine types.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




References
-
- Lo SW, et al. Pneumococcal lineages associated with serotype replacement and antibiotic resistance in childhood invasive pneumococcal disease in the post-PCV13 era: an international whole-genome sequencing study. Lancet Infect Dis. 2019;19:759–769. doi: 10.1016/s1473-3099(19)30297-x. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- 13/88/11/National Institute for Health Research (NIHR) Health Technology Assessment Programme, Antimicrobial Resistance Themed Call
- 13/88/11/National Institute for Health Research (NIHR) Health Technology Assessment Programme, Antimicrobial Resistance Themed Call
- 13/88/11/National Institute for Health Research (NIHR) Health Technology Assessment Programme, Antimicrobial Resistance Themed Call
- 13/88/11/National Institute for Health Research (NIHR) Health Technology Assessment Programme, Antimicrobial Resistance Themed Call
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

