Selective targeting of mu opioid receptors to primary cilia
- PMID: 38678559
- PMCID: PMC11257377
- DOI: 10.1016/j.celrep.2024.114164
Selective targeting of mu opioid receptors to primary cilia
Abstract
Opioid receptors are therapeutically important G protein-coupled receptors (GPCRs) with diverse neuromodulatory effects. The functional consequences of opioid receptor activation are known to depend on receptor location in the plasma membrane, but mechanisms mediating selective localization of receptors to any particular membrane domain remain elusive. Here, we demonstrate the targeting of the mu opioid receptor (MOR) to the primary cilium, a discrete microdomain of the somatic plasma membrane, both in vivo and in cultured cells. We further show that ciliary targeting is specific to MORs, requires a 17-residue sequence unique to the MOR cytoplasmic tail, and additionally requires the Tubby-like protein 3 (TULP3) ciliary adaptor protein. Our results reveal the potential for opioid receptors to undergo selective localization to the primary cilium. We propose that ciliary targeting is mediated through an elaboration of the recycling pathway, directed by a specific C-terminal recycling sequence in cis and requiring TULP3 in trans.
Keywords: CP: Cell biology; G protein-coupled receptor; MOR; TULP3; brain; mouse; neuronal; primary cilium; recycling.
Copyright © 2024 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures
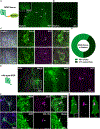

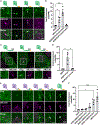
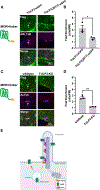
References
-
- Jimenez-Vargas NN, Gong J, Wisdom MJ, Jensen DD, Latorre R, Hegron A, Teng S, DiCello JJ, Rajasekhar P, Veldhuis NA, et al. (2020). Endosomal signaling of delta opioid receptors is an endogenous mechanism and therapeutic target for relief from inflammatory pain. Proc. Natl. Acad. Sci. USA 117, 15281–15292. 10.1073/pnas.2000500117. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

