DeepKEGG: a multi-omics data integration framework with biological insights for cancer recurrence prediction and biomarker discovery
- PMID: 38678587
- PMCID: PMC11056029
- DOI: 10.1093/bib/bbae185
DeepKEGG: a multi-omics data integration framework with biological insights for cancer recurrence prediction and biomarker discovery
Abstract
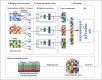
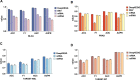
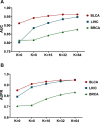
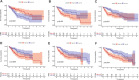
Deep learning-based multi-omics data integration methods have the capability to reveal the mechanisms of cancer development, discover cancer biomarkers and identify pathogenic targets. However, current methods ignore the potential correlations between samples in integrating multi-omics data. In addition, providing accurate biological explanations still poses significant challenges due to the complexity of deep learning models. Therefore, there is an urgent need for a deep learning-based multi-omics integration method to explore the potential correlations between samples and provide model interpretability. Herein, we propose a novel interpretable multi-omics data integration method (DeepKEGG) for cancer recurrence prediction and biomarker discovery. In DeepKEGG, a biological hierarchical module is designed for local connections of neuron nodes and model interpretability based on the biological relationship between genes/miRNAs and pathways. In addition, a pathway self-attention module is constructed to explore the correlation between different samples and generate the potential pathway feature representation for enhancing the prediction performance of the model. Lastly, an attribution-based feature importance calculation method is utilized to discover biomarkers related to cancer recurrence and provide a biological interpretation of the model. Experimental results demonstrate that DeepKEGG outperforms other state-of-the-art methods in 5-fold cross validation. Furthermore, case studies also indicate that DeepKEGG serves as an effective tool for biomarker discovery. The code is available at https://github.com/lanbiolab/DeepKEGG.
Keywords: cancer recurrence prediction; interpretability of deep learning; multi-omics data integration; self-attention mechanism.
© The Author(s) 2024. Published by Oxford University Press.
Figures


Similar articles
-
MORE: a multi-omics data-driven hypergraph integration network for biomedical data classification and biomarker identification.Brief Bioinform. 2024 Nov 22;26(1):bbae658. doi: 10.1093/bib/bbae658. Brief Bioinform. 2024. PMID: 39692449 Free PMC article.
-
Subtype-MGTP: a cancer subtype identification framework based on multi-omics translation.Bioinformatics. 2024 Jun 3;40(6):btae360. doi: 10.1093/bioinformatics/btae360. Bioinformatics. 2024. PMID: 38857453 Free PMC article.
-
Multi-omics integration method based on attention deep learning network for biomedical data classification.Comput Methods Programs Biomed. 2023 Apr;231:107377. doi: 10.1016/j.cmpb.2023.107377. Epub 2023 Jan 27. Comput Methods Programs Biomed. 2023. PMID: 36739624
-
A comprehensive review of machine learning techniques for multi-omics data integration: challenges and applications in precision oncology.Brief Funct Genomics. 2024 Sep 27;23(5):549-560. doi: 10.1093/bfgp/elae013. Brief Funct Genomics. 2024. PMID: 38600757 Review.
-
Multi-omics based artificial intelligence for cancer research.Adv Cancer Res. 2024;163:303-356. doi: 10.1016/bs.acr.2024.06.005. Epub 2024 Jul 9. Adv Cancer Res. 2024. PMID: 39271266 Review.
Cited by
-
PathX-CNN: An Enhanced Explainable Convolutional Neural Network for Survival Prediction and Pathway Analysis in Glioblastoma.bioRxiv [Preprint]. 2025 Jan 27:2025.01.24.634827. doi: 10.1101/2025.01.24.634827. bioRxiv. 2025. PMID: 39975150 Free PMC article. Preprint.
-
fuseMLR: an R package for integrative prediction modeling of multi-omics data.BMC Bioinformatics. 2025 Aug 26;26(1):221. doi: 10.1186/s12859-025-06248-4. BMC Bioinformatics. 2025. PMID: 40859122 Free PMC article.
-
Deciphering the molecular heterogeneity of intermediate- and (very-)high-risk non-muscle-invasive bladder cancer using multi-layered -omics studies.Front Oncol. 2024 Oct 21;14:1424293. doi: 10.3389/fonc.2024.1424293. eCollection 2024. Front Oncol. 2024. PMID: 39497708 Free PMC article.
-
Entropy measures for quantifying complexity in digital pathology and spatial omics.iScience. 2025 May 28;28(6):112765. doi: 10.1016/j.isci.2025.112765. eCollection 2025 Jun 20. iScience. 2025. PMID: 40546955 Free PMC article. Review.
-
DGHNN: a deep graph and hypergraph neural network for pan-cancer related gene prediction.Bioinformatics. 2025 Jul 1;41(7):btaf379. doi: 10.1093/bioinformatics/btaf379. Bioinformatics. 2025. PMID: 40580449 Free PMC article.
References
-
- Lan W, Wang J, Li M, et al. Computational approaches for prioritizing candidate disease genes based on PPI networks. Tsinghua Sci Technol 2015;20(5):500–12.
-
- Lan W, Dong Y, Chen Q, et al. KGANCDA: predicting circRNA-disease associations based on knowledge graph attention network. Brief Bioinform 2022;23(1):bbab494. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- 62072124/National Natural Science Foundation of China
- 2023JJG170006/Natural Science Foundation of Guangxi
- 2023JJ50117/Natural Science Foundation of Hunan Province
- 2022BZRC009/Natural Science and Technology Innovation Development Foundation of Guangxi University
- CAAIXSJLJJ-2022-022A/CAAI-Huawei MindSpore Open Fund

