Detection of SARS-CoV-2 B.1.1.529 (Omicron) variant by SYBR Green-based RT-qPCR
- PMID: 38680163
- PMCID: PMC11055497
- DOI: 10.1093/biomethods/bpae020
Detection of SARS-CoV-2 B.1.1.529 (Omicron) variant by SYBR Green-based RT-qPCR
Abstract
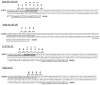
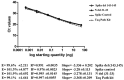
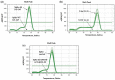
The coronavirus disease 2019 (COVID-19) pandemic is unceasingly spreading across the globe, and recently a highly transmissible Omicron SARS-CoV-2 variant (B.1.1.529) has been discovered in South Africa and Botswana. Rapid identification of this variant is essential for pandemic assessment and containment. However, variant identification is mainly being performed using expensive and time-consuming genomic sequencing. In this study, we propose an alternative RT-qPCR approach for the detection of the Omicron BA.1 variant using a low-cost and rapid SYBR Green method. We have designed specific primers to confirm the deletion mutations in the spike (S Δ143-145) and the nucleocapsid (N Δ31-33) which are characteristics of this variant. For the evaluation, we used 120 clinical samples from patients with PCR-confirmed SARS-CoV-2 infections, and displaying an S-gene target failure (SGTF) when using TaqPath COVID-19 kit (Thermo Fisher Scientific, Waltham, USA) that included the ORF1ab, S, and N gene targets. Our results showed that all the 120 samples harbored S Δ143-145 and N Δ31-33, which was further confirmed by whole-genome sequencing of 10 samples, thereby validating our SYBR Green-based protocol. This protocol can be easily implemented to rapidly confirm the diagnosis of the Omicron BA.1 variant in COVID-19 patients and prevent its spread among populations, especially in countries with high prevalence of SGTF profile.
Keywords: B.1.1.529; SARS-CoV2 RT-PCR; SGTF; variants of concern.
© The Author(s) 2024. Published by Oxford University Press.
Conflict of interest statement
The authors declare no competing interests.
Figures

Similar articles
-
A rapid and low-cost protocol for the detection of B.1.1.7 lineage of SARS-CoV-2 by using SYBR Green-based RT-qPCR.Mol Biol Rep. 2021 Nov;48(11):7243-7249. doi: 10.1007/s11033-021-06717-y. Epub 2021 Oct 6. Mol Biol Rep. 2021. PMID: 34613565 Free PMC article.
-
S-Gene Target Failure as an Effective Tool for Tracking the Emergence of Dominant SARS-CoV-2 Variants in Switzerland and Liechtenstein, Including Alpha, Delta, and Omicron BA.1, BA.2, and BA.4/BA.5.Microorganisms. 2024 Feb 3;12(2):321. doi: 10.3390/microorganisms12020321. Microorganisms. 2024. PMID: 38399725 Free PMC article.
-
Combined Use of RT-qPCR and NGS for Identification and Surveillance of SARS-CoV-2 Variants of Concern in Residual Clinical Laboratory Samples in Miami-Dade County, Florida.Viruses. 2023 Feb 21;15(3):593. doi: 10.3390/v15030593. Viruses. 2023. PMID: 36992302 Free PMC article.
-
The Omicron variant of concern: The genomics, diagnostics, and clinical characteristics in children.Front Pediatr. 2022 Aug 2;10:898463. doi: 10.3389/fped.2022.898463. eCollection 2022. Front Pediatr. 2022. PMID: 35983081 Free PMC article. Review.
-
The emergence of SARS-CoV-2 Omicron subvariants: current situation and future trends.Infez Med. 2022 Dec 1;30(4):480-494. doi: 10.53854/liim-3004-2. eCollection 2022. Infez Med. 2022. PMID: 36482957 Free PMC article. Review.
Cited by
-
A Smart Single-Loop-Mediated Isothermal Amplification Facilitates Flexible SNP Probe Design for On-Site Rapid Differentiation of SARS-CoV-2 Omicron Variants.Adv Sci (Weinh). 2025 Jul;12(26):e2502708. doi: 10.1002/advs.202502708. Epub 2025 Apr 1. Adv Sci (Weinh). 2025. PMID: 40167300 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Miscellaneous
