SARS-CoV-2 NSP5 antagonizes MHC II expression by subverting histone deacetylase 2
- PMID: 38682259
- PMCID: PMC11166459
- DOI: 10.1242/jcs.262172
SARS-CoV-2 NSP5 antagonizes MHC II expression by subverting histone deacetylase 2
Abstract
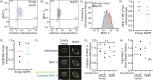
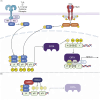
SARS-CoV-2 interferes with antigen presentation by downregulating major histocompatibility complex (MHC) II on antigen-presenting cells, but the mechanism mediating this process is unelucidated. Herein, analysis of protein and gene expression in human antigen-presenting cells reveals that MHC II is downregulated by the SARS-CoV-2 main protease, NSP5. This suppression of MHC II expression occurs via decreased expression of the MHC II regulatory protein CIITA. CIITA downregulation is independent of the proteolytic activity of NSP5, and rather, NSP5 delivers HDAC2 to the transcription factor IRF3 at an IRF-binding site within the CIITA promoter. Here, HDAC2 deacetylates and inactivates the CIITA promoter. This loss of CIITA expression prevents further expression of MHC II, with this suppression alleviated by ectopic expression of CIITA or knockdown of HDAC2. These results identify a mechanism by which SARS-CoV-2 limits MHC II expression, thereby delaying or weakening the subsequent adaptive immune response.
Keywords: Antigen presentation; CIITA; Dendritic cells; IRF3; MHC II; Macrophages; NSP5; SARS-CoV-2.
© 2024. Published by The Company of Biologists Ltd.
Conflict of interest statement
Competing interests The authors declare no competing or financial interests.
Figures





Similar articles
-
HDAC2 deacetylates class II transactivator and suppresses its activity in macrophages and smooth muscle cells.J Mol Cell Cardiol. 2009 Mar;46(3):292-9. doi: 10.1016/j.yjmcc.2008.10.023. Epub 2008 Nov 7. J Mol Cell Cardiol. 2009. PMID: 19041327
-
Histone deacetylase 1/mSin3A disrupts gamma interferon-induced CIITA function and major histocompatibility complex class II enhanceosome formation.Mol Cell Biol. 2003 May;23(9):3091-102. doi: 10.1128/MCB.23.9.3091-3102.2003. Mol Cell Biol. 2003. PMID: 12697811 Free PMC article.
-
Kaposi's Sarcoma-Associated Herpesvirus Latency-Associated Nuclear Antigen Inhibits Major Histocompatibility Complex Class II Expression by Disrupting Enhanceosome Assembly through Binding with the Regulatory Factor X Complex.J Virol. 2015 May;89(10):5536-56. doi: 10.1128/JVI.03713-14. Epub 2015 Mar 4. J Virol. 2015. PMID: 25740990 Free PMC article.
-
Expression of MHC II genes.Curr Top Microbiol Immunol. 2005;290:147-70. doi: 10.1007/3-540-26363-2_7. Curr Top Microbiol Immunol. 2005. PMID: 16480042 Review.
-
The MHC Class II Transactivator CIITA: Not (Quite) the Odd-One-Out Anymore among NLR Proteins.Int J Mol Sci. 2021 Jan 22;22(3):1074. doi: 10.3390/ijms22031074. Int J Mol Sci. 2021. PMID: 33499042 Free PMC article. Review.
Cited by
-
Impact of SARS-CoV-2 Wuhan and Omicron Variant Proteins on Type I Interferon Response.Viruses. 2025 Apr 15;17(4):569. doi: 10.3390/v17040569. Viruses. 2025. PMID: 40285011 Free PMC article.
-
The surveillance of viral infections by the unconventional Type I NKT cell.Front Immunol. 2024 Sep 17;15:1472854. doi: 10.3389/fimmu.2024.1472854. eCollection 2024. Front Immunol. 2024. PMID: 39355244 Free PMC article. Review.
References
-
- Bensussen, A., Álvarez-Buylla, E. R. and Díaz, J. (2021). SARS-CoV-2 Nsp5 protein causes acute lung inflammation, a dynamical mathematical model. Front. Syst. Biol. 1, 10.3389/fsysb.2021.764155. 10.3389/fsysb.2021.764155 - DOI
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

