Zymolyase Treatment of Saccharomyces cerevisiae Affects Cellular Proteins and Degrades Tyrosyl-DNA Phosphodiesterase I
- PMID: 38682313
- PMCID: PMC11322624
- DOI: 10.1089/dna.2024.0062
Zymolyase Treatment of Saccharomyces cerevisiae Affects Cellular Proteins and Degrades Tyrosyl-DNA Phosphodiesterase I
Abstract
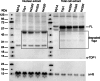
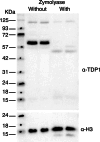
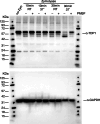
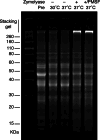
Saccharomyces cerevisiae is a genetically tractable, affordable, and extensively documented eukaryotic single-cell model organism. This budding yeast is amenable for the development of genetic and biochemical experiments and is frequently used to investigate the function, activity, and mechanism of mammalian proteins. However, yeast contains a cell wall that hinders select assays including organelle isolation. Lytic enzymes, with Zymolyase as the most effective and frequently used tool, are utilized to weaken the yeast cell wall resulting in yeast spheroplasts. Spheroplasts are easily lysed by, for example, osmotic-shock conditions to isolate yeast nuclei or mitochondria. However, during our studies of the DNA repair enzyme tyrosyl-DNA phosphodiesterase I (Tdp1), we encountered a negative effect of Zymolyase. We observed that Zymolyase treatment affected the steady-state protein levels of Tdp1. This was revealed by inconsistencies in technical and biological replicate lysates of plasmid-born galactose-induced expression of Tdp1. This off-target effect of Zymolyase is rarely discussed in articles and affects a select number of intracellular proteins, including transcription factors and assays such as chromatin immunoprecipitations. Following extensive troubleshooting, we concluded that the culprit is the Ser-protease, Zymolyase B, component of the Zymolyase enzyme mixture that causes the degradation of Tdp1. In this study, we report the protocols we have used, and our final protocol with an easy, affordable adaptation to any assay/protocol involving Zymolyase.
Keywords: TDP1; Zymolyase; budding yeast; nuclei; protein degradation; spheroplasts.
Figures

Similar articles
-
EORTC guidelines for the use of erythropoietic proteins in anaemic patients with cancer: 2006 update.Eur J Cancer. 2007 Jan;43(2):258-70. doi: 10.1016/j.ejca.2006.10.014. Epub 2006 Dec 19. Eur J Cancer. 2007. PMID: 17182241
-
Impact of residual disease as a prognostic factor for survival in women with advanced epithelial ovarian cancer after primary surgery.Cochrane Database Syst Rev. 2022 Sep 26;9(9):CD015048. doi: 10.1002/14651858.CD015048.pub2. Cochrane Database Syst Rev. 2022. PMID: 36161421 Free PMC article.
-
Drugs for preventing postoperative nausea and vomiting in adults after general anaesthesia: a network meta-analysis.Cochrane Database Syst Rev. 2020 Oct 19;10(10):CD012859. doi: 10.1002/14651858.CD012859.pub2. Cochrane Database Syst Rev. 2020. PMID: 33075160 Free PMC article.
-
Measures implemented in the school setting to contain the COVID-19 pandemic.Cochrane Database Syst Rev. 2022 Jan 17;1(1):CD015029. doi: 10.1002/14651858.CD015029. Cochrane Database Syst Rev. 2022. Update in: Cochrane Database Syst Rev. 2024 May 2;5:CD015029. doi: 10.1002/14651858.CD015029.pub2. PMID: 35037252 Free PMC article. Updated.
-
Interventions for promoting habitual exercise in people living with and beyond cancer.Cochrane Database Syst Rev. 2018 Sep 19;9(9):CD010192. doi: 10.1002/14651858.CD010192.pub3. Cochrane Database Syst Rev. 2018. PMID: 30229557 Free PMC article.
Cited by
-
Integrated Membrane Yeast Two-Hybrid System for the Analysis of Membrane Protein Complexes.Bio Protoc. 2025 Aug 20;15(16):e5418. doi: 10.21769/BioProtoc.5418. eCollection 2025 Aug 20. Bio Protoc. 2025. PMID: 40873472 Free PMC article.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

