Senescence of human pancreatic beta cells enhances functional maturation through chromatin reorganization and promotes interferon responsiveness
- PMID: 38682582
- PMCID: PMC11194086
- DOI: 10.1093/nar/gkae313
Senescence of human pancreatic beta cells enhances functional maturation through chromatin reorganization and promotes interferon responsiveness
Erratum in
-
Correction to 'Senescence of human pancreatic beta cells enhances functional maturation through chromatin reorganization and promotes interferon responsiveness'.Nucleic Acids Res. 2024 Jun 24;52(11):6732. doi: 10.1093/nar/gkae466. Nucleic Acids Res. 2024. PMID: 38801078 Free PMC article. No abstract available.
Abstract
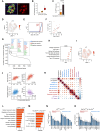
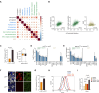
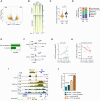
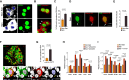
Senescent cells can influence the function of tissues in which they reside, and their propensity for disease. A portion of adult human pancreatic beta cells express the senescence marker p16, yet it is unclear whether they are in a senescent state, and how this affects insulin secretion. We analyzed single-cell transcriptome datasets of adult human beta cells, and found that p16-positive cells express senescence gene signatures, as well as elevated levels of beta-cell maturation genes, consistent with enhanced functionality. Senescent human beta-like cells in culture undergo chromatin reorganization that leads to activation of enhancers regulating functional maturation genes and acquisition of glucose-stimulated insulin secretion capacity. Strikingly, Interferon-stimulated genes are elevated in senescent human beta cells, but genes encoding senescence-associated secretory phenotype (SASP) cytokines are not. Senescent beta cells in culture and in human tissue show elevated levels of cytoplasmic DNA, contributing to their increased interferon responsiveness. Human beta-cell senescence thus involves chromatin-driven upregulation of a functional-maturation program, and increased responsiveness of interferon-stimulated genes, changes that could increase both insulin secretion and immune reactivity.
© The Author(s) 2024. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures



References
-
- Gorgoulis V., Adams P.D., Alimonti A., Bennett D.C., Bischof O., Bishop C., Campisi J., Collado M., Evangelou K., Ferbeyre G. et al. . Cellular senescence: defining a path forward. Cell. 2019; 179:813–827. - PubMed
-
- Olan I., Narita M. Senescence: an identity crisis originating from deep within the nucleus. Annu. Rev. Cell Dev. Biol. 2022; 38:219–239. - PubMed
MeSH terms
Substances
Grants and funding
- 65BX18MNIB/British Council BIRAX Program
- Stichting Onderzoek Nederland
- 3-SRA-2024-1479-S-B/Juvenile Diabetes Research Fund
- R01 DK133442/DK/NIDDK NIH HHS/United States
- Deutsches Krebsforschungszentrum
- U01 DK134995/DK/NIDDK NIH HHS/United States
- C9545/A29580/CRUK_/Cancer Research UK/United Kingdom
- Walter and Greta Stiel Chair and Research
- DKFZ
- U01 DK135001/DK/NIDDK NIH HHS/United States
- 0004062/Ministry of Science, Technology and Space
- C9545/A29580/Cancer Research UK Cambridge Institute
- Woll Sisters and Brothers Chair in Cardiovascular Diseases
- 1245/16/Israel Science Foundation Legacy Heritage Program
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

