Oxidative Stress-Mediated RUNX3 Mislocalization Occurs Via Jun Activation Domain-Binding Protein 1 and Histone Modification
- PMID: 38683453
- PMCID: PMC11645303
- DOI: 10.1007/s12010-024-04944-0
Oxidative Stress-Mediated RUNX3 Mislocalization Occurs Via Jun Activation Domain-Binding Protein 1 and Histone Modification
Abstract
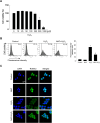
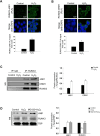
Runt domain transcription factor 3 (RUNX3) suppresses many different cancer types and is disabled by mutations, epigenetic repression, or cytoplasmic mislocalization. In this study, we investigated whether oxidative stress is associated with RUNX3 accumulation from the nucleus to the cytoplasm in terms of histone modification. Oxidative stress elevated histone deacetylase (HDAC) level and lowered that of histone acetyltransferase. In addition, oxidative stress decreased the expression of mixed lineage leukemia (MLL), a histone methyltransferase, but increased the expression of euchromatic histone-lysine N-methyltransferase 2 (EHMT2/G9a), which is also a histone methyltransferase. Moreover, oxidative stress-induced RUNX3 phosphorylation, Src activation, and Jun activation domain-binding protein 1 (JAB1) expression were inhibited by knockdown of HDAC and G9a, restoring the nuclear localization of RUNX3 under oxidative stress. Cytoplasmic RUNX3 localization was followed by oxidative stress-induced histone modification, activated Src along with RUNX3 phosphorylation, and induction of JAB1, resulting in RUNX3 inactivation.
Keywords: Cytoplasmic localization; Histone modification; Jun activation domain-binding protein 1; Oxidative stress; Runt domain transcription factor 3.
© 2024. The Author(s).
Conflict of interest statement
Declarations. Ethical Approval: Not applicable. Consent to Participate: Not applicable. Consent to Publication: Not applicable. Conflict of Interest: The authors declare no conflict of interest.
Figures





Similar articles
-
Cytoplasmic Localization of RUNX3 via Histone Deacetylase-Mediated SRC Expression in Oxidative-Stressed Colon Cancer Cells.J Cell Physiol. 2017 Jul;232(7):1914-1921. doi: 10.1002/jcp.25746. Epub 2017 Feb 21. J Cell Physiol. 2017. PMID: 27990641
-
Jab1/CSN5 induces the cytoplasmic localization and degradation of RUNX3.J Cell Biochem. 2009 Jun 1;107(3):557-65. doi: 10.1002/jcb.22157. J Cell Biochem. 2009. PMID: 19350572
-
Hypoxic silencing of tumor suppressor RUNX3 by histone modification in gastric cancer cells.Oncogene. 2009 Jan 15;28(2):184-94. doi: 10.1038/onc.2008.377. Epub 2008 Oct 13. Oncogene. 2009. PMID: 18850007
-
Control of RUNX3 by histone methyltransferases.J Cell Biochem. 2011 Feb;112(2):394-400. doi: 10.1002/jcb.22969. J Cell Biochem. 2011. PMID: 21268059 Review.
-
Prognostic role of c-Jun activation domain-binding protein-1 in cancer: A systematic review and meta-analysis.J Cell Mol Med. 2021 Mar;25(6):2750-2763. doi: 10.1111/jcmm.16334. Epub 2021 Feb 7. J Cell Mol Med. 2021. PMID: 33550701 Free PMC article.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

