Unravelling the role of the group 6 soluble di-iron monooxygenase (SDIMO) SmoABCD in alkane metabolism and chlorinated alkane degradation
- PMID: 38683670
- PMCID: PMC11057499
- DOI: 10.1111/1751-7915.14453
Unravelling the role of the group 6 soluble di-iron monooxygenase (SDIMO) SmoABCD in alkane metabolism and chlorinated alkane degradation
Abstract
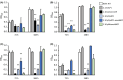
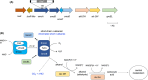
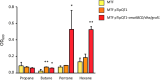
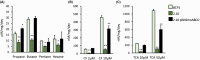
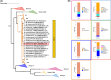
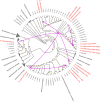
Soluble di-iron monooxygenases (SDIMOs) are multi-component enzymes catalysing the oxidation of various substrates. These enzymes are characterized by high sequence and functional diversity that is still not well understood despite their key role in biotechnological processes including contaminant biodegradation. In this study, we analysed a mutant of Rhodoccocus aetherivorans BCP1 (BCP1-2.10) characterized by a transposon insertion in the gene smoA encoding the alpha subunit of the plasmid-located SDIMO SmoABCD. The mutant BCP1-2.10 showed a reduced capacity to grow on propane, lost the ability to grow on butane, pentane and n-hexane and was heavily impaired in the capacity to degrade chloroform and trichloroethane. The expression of the additional SDIMO prmABCD in BCP1-2.10 probably allowed the mutant to partially grow on propane and to degrade it, to some extent, together with the other short-chain n-alkanes. The complementation of the mutant, conducted by introducing smoABCD in the genome as a single copy under a constitutive promoter or within a plasmid under a thiostreptone-inducible promoter, allowed the recovery of the alkanotrophic phenotype as well as the capacity to degrade chlorinated n-alkanes. The heterologous expression of smoABCD allowed a non-alkanotrophic Rhodococcus strain to grow on pentane and n-hexane when the gene cluster was introduced together with the downstream genes encoding alcohol and aldehyde dehydrogenases and a GroEL chaperon. BCP1 smoA gene was shown to belong to the group 6 SDIMOs, which is a rare group of monooxygenases mostly present in Mycobacterium genus and in a few Rhodococcus strains. SmoABCD originally evolved in Mycobacterium and was then acquired by Rhodococcus through horizontal gene transfer events. This work extends the knowledge of the biotechnologically relevant SDIMOs by providing functional and evolutionary insights into a group 6 SDIMO in Rhodococcus and demonstrating its key role in the metabolism of short-chain alkanes and degradation of chlorinated n-alkanes.
© 2024 The Authors. Microbial Biotechnology published by John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
Growth of Rhodococcus sp. strain BCP1 on gaseous n-alkanes: new metabolic insights and transcriptional analysis of two soluble di-iron monooxygenase genes.Front Microbiol. 2015 May 12;6:393. doi: 10.3389/fmicb.2015.00393. eCollection 2015. Front Microbiol. 2015. PMID: 26029173 Free PMC article.
-
Horizontal Gene Transfer of Genes Encoding Copper-Containing Membrane-Bound Monooxygenase (CuMMO) and Soluble Di-iron Monooxygenase (SDIMO) in Ethane- and Propane-Oxidizing Rhodococcus Bacteria.Appl Environ Microbiol. 2021 Jun 25;87(14):e0022721. doi: 10.1128/AEM.00227-21. Epub 2021 Jun 25. Appl Environ Microbiol. 2021. PMID: 33962978 Free PMC article.
-
Analyses of both the alkB gene transcriptional start site and alkB promoter-inducing properties of Rhodococcus sp. strain BCP1 grown on n-alkanes.Appl Environ Microbiol. 2011 Mar;77(5):1619-27. doi: 10.1128/AEM.01987-10. Epub 2010 Dec 30. Appl Environ Microbiol. 2011. PMID: 21193665 Free PMC article.
-
Phylogenetic and Functional Diversity of Soluble Di-Iron Monooxygenases.Environ Microbiol. 2025 Feb;27(2):e70050. doi: 10.1111/1462-2920.70050. Environ Microbiol. 2025. PMID: 39947201 Free PMC article. Review.
-
Genetics of alkane oxidation by Pseudomonas oleovorans.Biodegradation. 1994 Dec;5(3-4):161-74. doi: 10.1007/BF00696457. Biodegradation. 1994. PMID: 7532480 Review.
Cited by
-
Resistant Rhodococcus for Biodegradation of Diesel Fuel at High Concentration and Low Temperature.Microorganisms. 2024 Dec 17;12(12):2605. doi: 10.3390/microorganisms12122605. Microorganisms. 2024. PMID: 39770807 Free PMC article.
-
The Lipid- and Polysaccharide-Rich Extracellular Polymeric Substances of Rhodococcus Support Biofilm Formation and Protection from Toxic Hydrocarbons.Polymers (Basel). 2025 Jul 10;17(14):1912. doi: 10.3390/polym17141912. Polymers (Basel). 2025. PMID: 40732790 Free PMC article.
-
Generalization of Classification of AlkB Family Alkane Monooxygenases from Rhodococcus (sensu lato) Group Based on Phylogenetic Analysis and Genomic Context Comparison.Int J Mol Sci. 2025 Feb 17;26(4):1713. doi: 10.3390/ijms26041713. Int J Mol Sci. 2025. PMID: 40004181 Free PMC article.
References
-
- Cappelletti, M. , Fedi, S. , Frascari, D. , Ohtake, H. , Turner, R.J. & Zannoni, D. (2011) Analyses of both the alkB gene transcriptional start site and alkB promoter‐inducing properties of Rhodococcus sp. strain BCP1 grown on n‐alkanes. Applied and Environmental Microbiology, 77(5), 1619–1627. - PMC - PubMed
-
- Cappelletti, M. , Fedi, S. , Zampolli, J. , Di Canito, A. , D'Ursi, P. , Orro, A. et al. (2016) Phenotype microarray analysis may unravel genetic determinants of the stress response by Rhodococcus aetherivorans BCP1 and Rhodococcus opacus R7. Research in Microbiology, 167(9–10), 766–773. Available from: 10.1016/j.resmic.2016.06.008 - DOI - PubMed
-
- Cappelletti, M. , Fedi, S. & Zannoni, D. (2019) Degradation of alkanes in Rhodococcus . In: Alvarez, H. (Ed.) Biology of Rhodococcus, Vol. 16. Cham, Switzerland: Springer International Publishing, pp. 137–171. Available from: 10.1007/978-3-030-11461-9_6 - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

