Structural characterization of two nanobodies targeting the ligand-binding pocket of human Arc
- PMID: 38683783
- PMCID: PMC11057775
- DOI: 10.1371/journal.pone.0300453
Structural characterization of two nanobodies targeting the ligand-binding pocket of human Arc
Erratum in
-
Correction: Structural characterization of two nanobodies targeting the ligand-binding pocket of human Arc.PLoS One. 2025 Mar 25;20(3):e0321004. doi: 10.1371/journal.pone.0321004. eCollection 2025. PLoS One. 2025. PMID: 40131924 Free PMC article.
Abstract
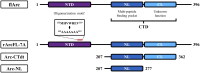
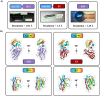
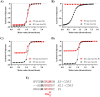
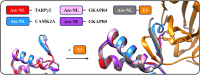
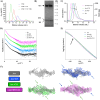
The activity-regulated cytoskeleton-associated protein (Arc) is a complex regulator of synaptic plasticity in glutamatergic neurons. Understanding its molecular function is key to elucidate the neurobiology of memory and learning, stress regulation, and multiple neurological and psychiatric diseases. The recent development of anti-Arc nanobodies has promoted the characterization of the molecular structure and function of Arc. This study aimed to validate two anti-Arc nanobodies, E5 and H11, as selective modulators of the human Arc N-lobe (Arc-NL), a domain that mediates several molecular functions of Arc through its peptide ligand binding site. The structural characteristics of recombinant Arc-NL-nanobody complexes were solved at atomic resolution using X-ray crystallography. Both anti-Arc nanobodies bind specifically to the multi-peptide binding site of Arc-NL. Isothermal titration calorimetry showed that the Arc-NL-nanobody interactions occur at nanomolar affinity, and that the nanobodies can displace a TARPγ2-derived peptide from the binding site. Thus, both anti-Arc-NL nanobodies could be used as competitive inhibitors of endogenous Arc ligands. Differences in the CDR3 loops between the two nanobodies indicate that the spectrum of short linear motifs recognized by the Arc-NL should be expanded. We provide a robust biochemical background to support the use of anti-Arc nanobodies in attempts to target Arc-dependent synaptic plasticity. Function-blocking anti-Arc nanobodies could eventually help unravel the complex neurobiology of synaptic plasticity and allow to develop diagnostic and treatment tools.
Copyright: © 2024 Godoy Muñoz et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


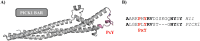
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

