Reconstruction Set Test (RESET): A computationally efficient method for single sample gene set testing based on randomized reduced rank reconstruction error
- PMID: 38683883
- PMCID: PMC11081506
- DOI: 10.1371/journal.pcbi.1012084
Reconstruction Set Test (RESET): A computationally efficient method for single sample gene set testing based on randomized reduced rank reconstruction error
Abstract
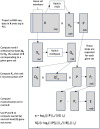
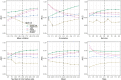
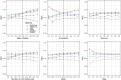
We have developed a new, and analytically novel, single sample gene set testing method called Reconstruction Set Test (RESET). RESET quantifies gene set importance based on the ability of set genes to reconstruct values for all measured genes. RESET is realized using a computationally efficient randomized reduced rank reconstruction algorithm (available via the RESET R package on CRAN) that can effectively detect patterns of differential abundance and differential correlation for self-contained and competitive scenarios. As demonstrated using real and simulated scRNA-seq data, RESET provides superior performance at a lower computational cost relative to other single sample approaches.
Copyright: © 2024 H. Robert Frost. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures




Update of
-
Reconstruction Set Test (RESET): a computationally efficient method for single sample gene set testing based on randomized reduced rank reconstruction error.bioRxiv [Preprint]. 2023 Apr 20:2023.04.03.535366. doi: 10.1101/2023.04.03.535366. bioRxiv. 2023. Update in: PLoS Comput Biol. 2024 Apr 29;20(4):e1012084. doi: 10.1371/journal.pcbi.1012084. PMID: 37066315 Free PMC article. Updated. Preprint.
Similar articles
-
Reconstruction Set Test (RESET): a computationally efficient method for single sample gene set testing based on randomized reduced rank reconstruction error.bioRxiv [Preprint]. 2023 Apr 20:2023.04.03.535366. doi: 10.1101/2023.04.03.535366. bioRxiv. 2023. Update in: PLoS Comput Biol. 2024 Apr 29;20(4):e1012084. doi: 10.1371/journal.pcbi.1012084. PMID: 37066315 Free PMC article. Updated. Preprint.
-
Nonlinear Network Reconstruction from Gene Expression Data Using Marginal Dependencies Measured by DCOL.PLoS One. 2016 Jul 5;11(7):e0158247. doi: 10.1371/journal.pone.0158247. eCollection 2016. PLoS One. 2016. PMID: 27380516 Free PMC article.
-
Improving Gene-Set Enrichment Analysis of RNA-Seq Data with Small Replicates.PLoS One. 2016 Nov 9;11(11):e0165919. doi: 10.1371/journal.pone.0165919. eCollection 2016. PLoS One. 2016. PMID: 27829002 Free PMC article.
-
Variance-adjusted Mahalanobis (VAM): a fast and accurate method for cell-specific gene set scoring.Nucleic Acids Res. 2020 Sep 18;48(16):e94. doi: 10.1093/nar/gkaa582. Nucleic Acids Res. 2020. PMID: 32633778 Free PMC article.
-
SILGGM: An extensive R package for efficient statistical inference in large-scale gene networks.PLoS Comput Biol. 2018 Aug 13;14(8):e1006369. doi: 10.1371/journal.pcbi.1006369. eCollection 2018 Aug. PLoS Comput Biol. 2018. PMID: 30102702 Free PMC article.
Cited by
-
GRPa-PRS: A risk stratification method to identify genetically-regulated pathways in polygenic diseases.medRxiv [Preprint]. 2024 Jul 5:2023.06.19.23291621. doi: 10.1101/2023.06.19.23291621. medRxiv. 2024. PMID: 37425929 Free PMC article. Preprint.
-
Leveraging cell type-specificity for gene set analysis of single cell transcriptomics.bioRxiv [Preprint]. 2024 Sep 27:2024.09.25.615040. doi: 10.1101/2024.09.25.615040. bioRxiv. 2024. PMID: 39386631 Free PMC article. Preprint.
-
Gene set optimization for single cell transcriptomics.Comput Intell Methods Bioinform Biostat. 2025;15276:183-195. doi: 10.1007/978-3-031-89704-7_14. Epub 2025 May 15. Comput Intell Methods Bioinform Biostat. 2025. PMID: 40487192 Free PMC article.
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

