HA-1-targeted T-cell receptor T-cell therapy for recurrent leukemia after hematopoietic stem cell transplantation
- PMID: 38683966
- PMCID: PMC11406181
- DOI: 10.1182/blood.2024024105
HA-1-targeted T-cell receptor T-cell therapy for recurrent leukemia after hematopoietic stem cell transplantation
Abstract
Relapse is the leading cause of death after allogeneic hematopoietic stem cell transplantation (HCT) for leukemia. T cells engineered by gene transfer to express T cell receptors (TCR; TCR-T) specific for hematopoietic-restricted minor histocompatibility (H) antigens may provide a potent selective antileukemic effect post-HCT. We conducted a phase 1 clinical trial using a novel TCR-T product targeting the minor H antigen, HA-1, to treat or consolidate treatment of persistent or recurrent leukemia and myeloid neoplasms. The primary objective was to evaluate the feasibility and safety of administration of HA-1 TCR-T after HCT. CD8+ and CD4+ T cells expressing the HA-1 TCR and a CD8 coreceptor were successfully manufactured from HA-1-disparate HCT donors. One or more infusions of HA-1 TCR-T following lymphodepleting chemotherapy were administered to 9 HCT recipients who had developed disease recurrence after HCT. TCR-T cells expanded and persisted in vivo after adoptive transfer. No dose-limiting toxicities occurred. Although the study was not designed to assess efficacy, 4 patients achieved or maintained complete remissions following lymphodepletion and HA-1 TCR-T, with 1 patient still in remission at >2 years. Single-cell RNA sequencing of relapsing/progressive leukemia after TCR-T therapy identified upregulated molecules associated with T-cell dysfunction or cancer cell survival. HA-1 TCR-T therapy appears feasible and safe and shows preliminary signals of efficacy. This clinical trial was registered at ClinicalTrials.gov as #NCT03326921.
© 2024 American Society of Hematology. Published by Elsevier Inc. All rights are reserved, including those for text and data mining, AI training, and similar technologies.
Conflict of interest statement
Conflict-of-interest disclosure: M. Bleakley and R.G.D. are coinventors on a patent describing HA-1 T-cell receptor therapy that was previously licensed to ElevateBio and has recently been licensed to PromiCell Therapeutics (TCRS SPECIFIC FOR MINOR H ANTIGEN HA-1 AND USES THEREOF; US 10,538,577 B2 Jan 21 2020; Inventors M. Bleakley, Robson Dossa, Daniel Sommermeyer). M. Bleakley received funding from HighPassBio, an ElevateBio portfolio company, and was a scientific advisory board member and had equity in HighPassBio. HighPassBio provided funding for the clinical trial on which E.F.K. was the principal investigator. At the time of the research, A.E.D. was an employee of ElevateBio Technologies and is currently an owner of stock in ElevateBio LLC. E.F.K. served on an advisory board for TScan Therapeutics. R.D.C. has received research funding from Amgen, Kite/Gilead, Incyte, Merck, Pfizer, Servier, and Vanda Pharmaceuticals; consultancy/honoraria from Amgen, Jazz, Kite/Gilead, and Pfizer; and membership on a board or advisory committee for Autolus and PeproMene Bio. The remaining authors declare no competing financial interests.
The current affiliation of R.G.D. is Arovella Therapeutics, Melbourne, VIC, Australia.
The current affiliation of M. Bar is Bristol-Myers Squibb, Summit, NJ.
Figures


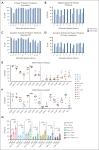
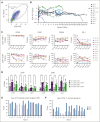


Comment in
-
TCR targeting hematopoiesis to cure leukemia.Blood. 2024 Sep 5;144(10):1031-1033. doi: 10.1182/blood.2024024972. Blood. 2024. PMID: 39235801 No abstract available.
References
-
- Fiorenza S, Turtle CJ. CAR-T cell therapy for acute myeloid leukemia: preclinical rationale, current clinical progress, and barriers to success. BioDrugs. 2021;35(3):281–302. - PubMed
-
- Bleakley M, Riddell SR. Molecules and mechanisms of the graft-versus-leukaemia effect. Nat Rev Cancer. 2004;4(5):371–380. - PubMed
Publication types
MeSH terms
Substances
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

