scTPC: a novel semisupervised deep clustering model for scRNA-seq data
- PMID: 38684178
- PMCID: PMC11091743
- DOI: 10.1093/bioinformatics/btae293
scTPC: a novel semisupervised deep clustering model for scRNA-seq data
Abstract
Motivation: Continuous advancements in single-cell RNA sequencing (scRNA-seq) technology have enabled researchers to further explore the study of cell heterogeneity, trajectory inference, identification of rare cell types, and neurology. Accurate scRNA-seq data clustering is crucial in single-cell sequencing data analysis. However, the high dimensionality, sparsity, and presence of "false" zero values in the data can pose challenges to clustering. Furthermore, current unsupervised clustering algorithms have not effectively leveraged prior biological knowledge, making cell clustering even more challenging.
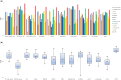
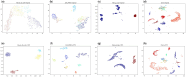
Results: This study investigates a semisupervised clustering model called scTPC, which integrates the triplet constraint, pairwise constraint, and cross-entropy constraint based on deep learning. Specifically, the model begins by pretraining a denoising autoencoder based on a zero-inflated negative binomial distribution. Deep clustering is then performed in the learned latent feature space using triplet constraints and pairwise constraints generated from partial labeled cells. Finally, to address imbalanced cell-type datasets, a weighted cross-entropy loss is introduced to optimize the model. A series of experimental results on 10 real scRNA-seq datasets and five simulated datasets demonstrate that scTPC achieves accurate clustering with a well-designed framework.
Availability and implementation: scTPC is a Python-based algorithm, and the code is available from https://github.com/LF-Yang/Code or https://zenodo.org/records/10951780.
© The Author(s) 2024. Published by Oxford University Press.
Conflict of interest statement
The authors declared that they have no conflicts of interest.
Figures






Similar articles
-
scBGEDA: deep single-cell clustering analysis via a dual denoising autoencoder with bipartite graph ensemble clustering.Bioinformatics. 2023 Feb 14;39(2):btad075. doi: 10.1093/bioinformatics/btad075. Bioinformatics. 2023. PMID: 36734596 Free PMC article.
-
scAMZI: attention-based deep autoencoder with zero-inflated layer for clustering scRNA-seq data.BMC Genomics. 2025 Apr 7;26(1):350. doi: 10.1186/s12864-025-11511-2. BMC Genomics. 2025. PMID: 40197174 Free PMC article.
-
scDCCA: deep contrastive clustering for single-cell RNA-seq data based on auto-encoder network.Brief Bioinform. 2023 Jan 19;24(1):bbac625. doi: 10.1093/bib/bbac625. Brief Bioinform. 2023. PMID: 36631401
-
Machine learning and statistical methods for clustering single-cell RNA-sequencing data.Brief Bioinform. 2020 Jul 15;21(4):1209-1223. doi: 10.1093/bib/bbz063. Brief Bioinform. 2020. PMID: 31243426 Review.
-
Application of Deep Learning on Single-cell RNA Sequencing Data Analysis: A Review.Genomics Proteomics Bioinformatics. 2022 Oct;20(5):814-835. doi: 10.1016/j.gpb.2022.11.011. Epub 2022 Dec 14. Genomics Proteomics Bioinformatics. 2022. PMID: 36528240 Free PMC article. Review.
Cited by
-
A robust multi-scale clustering framework for single-cell RNA-seq data analysis.Sci Rep. 2025 May 27;15(1):18543. doi: 10.1038/s41598-025-03603-6. Sci Rep. 2025. PMID: 40425750 Free PMC article.
-
SpaGIC: graph-informed clustering in spatial transcriptomics via self-supervised contrastive learning.Brief Bioinform. 2024 Sep 23;25(6):bbae578. doi: 10.1093/bib/bbae578. Brief Bioinform. 2024. PMID: 39541189 Free PMC article.
-
scSAMAC: saliency-adjusted masking induced attention contrastive learning for single-cell clustering.Brief Bioinform. 2025 Mar 4;26(2):bbaf128. doi: 10.1093/bib/bbaf128. Brief Bioinform. 2025. PMID: 40131310 Free PMC article.
-
SCassist: an AI based workflow assistant for single-cell analysis.Bioinformatics. 2025 Aug 2;41(8):btaf402. doi: 10.1093/bioinformatics/btaf402. Bioinformatics. 2025. PMID: 40650988 Free PMC article.
-
scRECL: representative ensembles with contrastive learning for scRNA-seq data clustering analysis.Brief Bioinform. 2025 Jul 2;26(4):bbaf346. doi: 10.1093/bib/bbaf346. Brief Bioinform. 2025. PMID: 40671174 Free PMC article.
References
-
- Basu S, Davidson I, Wagstaff K.. Constrained Clustering: Advances in Algorithms, Theory, and Applications. New York: CRC Press, 2008.
-
- Chen L, He Q, Zhai Y. et al. Single-cell RNA-seq data semi-supervised clustering and annotation via structural regularized domain adaptation. Bioinformatics 2021;37:775–84. - PubMed

