Sulfur oxidation and reduction are coupled to nitrogen fixation in the roots of the salt marsh foundation plant Spartina alterniflora
- PMID: 38684658
- PMCID: PMC11059160
- DOI: 10.1038/s41467-024-47646-1
Sulfur oxidation and reduction are coupled to nitrogen fixation in the roots of the salt marsh foundation plant Spartina alterniflora
Abstract
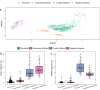
Heterotrophic activity, primarily driven by sulfate-reducing prokaryotes, has traditionally been linked to nitrogen fixation in the root zone of coastal marine plants, leaving the role of chemolithoautotrophy in this process unexplored. Here, we show that sulfur oxidation coupled to nitrogen fixation is a previously overlooked process providing nitrogen to coastal marine macrophytes. In this study, we recovered 239 metagenome-assembled genomes from a salt marsh dominated by the foundation plant Spartina alterniflora, including diazotrophic sulfate-reducing and sulfur-oxidizing bacteria. Abundant sulfur-oxidizing bacteria encode and highly express genes for carbon fixation (RuBisCO), nitrogen fixation (nifHDK) and sulfur oxidation (oxidative-dsrAB), especially in roots stressed by sulfidic and reduced sediment conditions. Stressed roots exhibited the highest rates of nitrogen fixation and expression level of sulfur oxidation and sulfate reduction genes. Close relatives of marine symbionts from the Candidatus Thiodiazotropha genus contributed ~30% and ~20% of all sulfur-oxidizing dsrA and nitrogen-fixing nifK transcripts in stressed roots, respectively. Based on these findings, we propose that the symbiosis between S. alterniflora and sulfur-oxidizing bacteria is key to ecosystem functioning of coastal salt marshes.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures





References
-
- Barbier EB, et al. The value of estuarine and coastal ecosystem services. Ecol. Monogr. 2011;81:169–193. doi: 10.1890/10-1510.1. - DOI
-
- Duarte CM, Losada IJ, Hendriks IE, Mazarrasa I, Marbà N. The role of coastal plant communities for climate change mitigation and adaptation. Nat. Clim. change. 2013;3:961–968. doi: 10.1038/nclimate1970. - DOI
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

