VC-resist glioblastoma cell state: vessel co-option as a key driver of chemoradiation resistance
- PMID: 38684700
- PMCID: PMC11058782
- DOI: 10.1038/s41467-024-47985-z
VC-resist glioblastoma cell state: vessel co-option as a key driver of chemoradiation resistance
Abstract
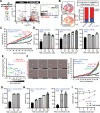
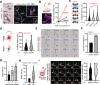
Glioblastoma (GBM) is a highly lethal type of cancer. GBM recurrence following chemoradiation is typically attributed to the regrowth of invasive and resistant cells. Therefore, there is a pressing need to gain a deeper understanding of the mechanisms underlying GBM resistance to chemoradiation and its ability to infiltrate. Using a combination of transcriptomic, proteomic, and phosphoproteomic analyses, longitudinal imaging, organotypic cultures, functional assays, animal studies, and clinical data analyses, we demonstrate that chemoradiation and brain vasculature induce cell transition to a functional state named VC-Resist (vessel co-opting and resistant cell state). This cell state is midway along the transcriptomic axis between proneural and mesenchymal GBM cells and is closer to the AC/MES1-like state. VC-Resist GBM cells are highly vessel co-opting, allowing significant infiltration into the surrounding brain tissue and homing to the perivascular niche, which in turn induces even more VC-Resist transition. The molecular and functional characteristics of this FGFR1-YAP1-dependent GBM cell state, including resistance to DNA damage, enrichment in the G2M phase, and induction of senescence/stemness pathways, contribute to its enhanced resistance to chemoradiation. These findings demonstrate how vessel co-option, perivascular niche, and GBM cell plasticity jointly drive resistance to therapy during GBM recurrence.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures








References
Publication types
MeSH terms
Substances
Grants and funding
- 805225/EC | EU Framework Programme for Research and Innovation H2020 | H2020 Priority Excellent Science | H2020 European Research Council (H2020 Excellent Science - European Research Council)
- NanoTheRad/Université Paris-Saclay (University of Paris-Saclay)
- ATIP-Avenir/Centre National de la Recherche Scientifique (National Center for Scientific Research)
- INCa-2021-1-PAIR-CEREB-01-1/Institut National Du Cancer (French National Cancer Institute)
- R01 LM013897/LM/NLM NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

