Mitochondrial complex I deficiency stratifies idiopathic Parkinson's disease
- PMID: 38684731
- PMCID: PMC11059185
- DOI: 10.1038/s41467-024-47867-4
Mitochondrial complex I deficiency stratifies idiopathic Parkinson's disease
Abstract
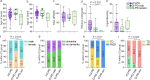
Idiopathic Parkinson's disease (iPD) is believed to have a heterogeneous pathophysiology, but molecular disease subtypes have not been identified. Here, we show that iPD can be stratified according to the severity of neuronal respiratory complex I (CI) deficiency, and identify two emerging disease subtypes with distinct molecular and clinical profiles. The CI deficient (CI-PD) subtype accounts for approximately a fourth of all cases, and is characterized by anatomically widespread neuronal CI deficiency, a distinct cell type-specific gene expression profile, increased load of neuronal mtDNA deletions, and a predilection for non-tremor dominant motor phenotypes. In contrast, the non-CI deficient (nCI-PD) subtype exhibits no evidence of mitochondrial impairment outside the dopaminergic substantia nigra and has a predilection for a tremor dominant phenotype. These findings constitute a step towards resolving the biological heterogeneity of iPD with implications for both mechanistic understanding and treatment strategies.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures





References
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

