Wnt/β-catenin signaling regulates amino acid metabolism through the suppression of CEBPA and FOXA1 in liver cancer cells
- PMID: 38684876
- PMCID: PMC11058205
- DOI: 10.1038/s42003-024-06202-9
Wnt/β-catenin signaling regulates amino acid metabolism through the suppression of CEBPA and FOXA1 in liver cancer cells
Abstract
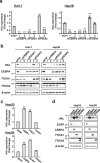
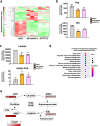
Deregulation of the Wnt/β-catenin pathway is associated with the development of human cancer including colorectal and liver cancer. Although we previously showed that histidine ammonia lyase (HAL) was transcriptionally reduced by the β-catenin/TCF complex in liver cancer cells, the mechanism(s) of its down-regulation by the complex remain to be clarified. In this study, we search for the transcription factor(s) regulating HAL, and identify CEBPA and FOXA1, two factors whose expression is suppressed by the knockdown of β-catenin or TCF7L2. In addition, RNA-seq analysis coupled with genome-wide mapping of CEBPA- and FOXA1-binding regions reveals that these two factors also increase the expression of arginase 1 (ARG1) that catalyzes the hydrolysis of arginine. Metabolome analysis discloses that activated Wnt signaling augments intracellular concentrations of histidine and arginine, and that the signal also increases the level of lactic acid suggesting the induction of the Warburg effect in liver cancer cells. Further analysis reveals that the levels of metabolites of the urea cycle and genes coding its related enzymes are also modulated by the Wnt signaling. These findings shed light on the altered cellular metabolism in the liver by the Wnt/β-catenin pathway through the suppression of liver-enriched transcription factors including CEBPA and FOXA1.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

