Cold-induced expression of a truncated adenylyl cyclase 3 acts as rheostat to brown fat function
- PMID: 38684889
- PMCID: PMC11971047
- DOI: 10.1038/s42255-024-01033-8
Cold-induced expression of a truncated adenylyl cyclase 3 acts as rheostat to brown fat function
Erratum in
-
Publisher Correction: Cold-induced expression of a truncated adenylyl cyclase 3 acts as rheostat to brown fat function.Nat Metab. 2025 Apr;7(4):855. doi: 10.1038/s42255-025-01292-z. Nat Metab. 2025. PMID: 40186076 Free PMC article. No abstract available.
Abstract
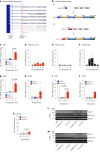
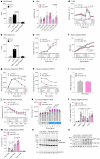
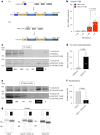
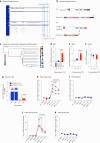
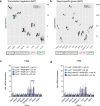
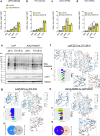
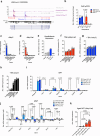
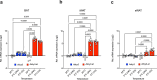
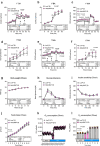
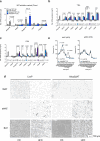
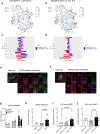
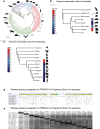
Promoting brown adipose tissue (BAT) activity innovatively targets obesity and metabolic disease. While thermogenic activation of BAT is well understood, the rheostatic regulation of BAT to avoid excessive energy dissipation remains ill-defined. Here, we demonstrate that adenylyl cyclase 3 (AC3) is key for BAT function. We identified a cold-inducible promoter that generates a 5' truncated AC3 mRNA isoform (Adcy3-at), whose expression is driven by a cold-induced, truncated isoform of PPARGC1A (PPARGC1A-AT). Male mice lacking Adcy3-at display increased energy expenditure and are resistant to obesity and ensuing metabolic imbalances. Mouse and human AC3-AT are retained in the endoplasmic reticulum, unable to translocate to the plasma membrane and lack enzymatic activity. AC3-AT interacts with AC3 and sequesters it in the endoplasmic reticulum, reducing the pool of adenylyl cyclases available for G-protein-mediated cAMP synthesis. Thus, AC3-AT acts as a cold-induced rheostat in BAT, limiting adverse consequences of cAMP activity during chronic BAT activation.
© 2024. The Author(s), under exclusive licence to Springer Nature Limited.
Conflict of interest statement
The authors declare no competing interests.
Figures





References
-
- Kusminski, C. M., Bickel, P. E. & Scherer, P. E. Targeting adipose tissue in the treatment of obesity-associated diabetes. Nat. Rev. Drug Discov.15, 639–660 (2016). - PubMed
-
- Cannon, B. & Nedergaard, J. Brown adipose tissue: function and physiological significance. Physiol. Rev.84, 277–359 (2004). - PubMed
-
- van Marken Lichtenbelt, W. D. et al. Cold-activated brown adipose tissue in healthy men. N. Engl. J. Med.360, 1500–1508 (2009). - PubMed
-
- Virtanen, K. A. et al. Functional brown adipose tissue in healthy adults. N. Engl. J. Med.360, 1518–1525 (2009). - PubMed
MeSH terms
Substances
Grants and funding
- 675014/EC | EC Seventh Framework Programm | FP7 Ideas: European Research Council (FP7-IDEAS-ERC - Specific Programme: "Ideas" Implementing the Seventh Framework Programme of the European Community for Research, Technological Development and Demonstration Activities (2007 to 2013))
- PROTEOFIT/EC | EC Seventh Framework Programm | FP7 Ideas: European Research Council (FP7-IDEAS-ERC - Specific Programme: "Ideas" Implementing the Seventh Framework Programme of the European Community for Research, Technological Development and Demonstration Activities (2007 to 2013))
- 33444/Novo Nordisk Fonden (Novo Nordisk Foundation)
- 28416/Novo Nordisk Fonden (Novo Nordisk Foundation)
- A/12/97620/Deutscher Akademischer Austauschdienst (German Academic Exchange Service)
- TRR333/1 (450149205)/Deutsche Forschungsgemeinschaft (German Research Foundation)
- SFB 1454 (432325352)/Deutsche Forschungsgemeinschaft (German Research Foundation)
- TRR83/Deutsche Forschungsgemeinschaft (German Research Foundation)
- SPP1926/Deutsche Forschungsgemeinschaft (German Research Foundation)
- SPP1726/Deutsche Forschungsgemeinschaft (German Research Foundation)
- FOR2743/Deutsche Forschungsgemeinschaft (German Research Foundation)
- SFB1123-B10/Deutsche Forschungsgemeinschaft (German Research Foundation)
- 676-2021/European Molecular Biology Organization (EMBO)
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

