Microbial communities colonising plastics during transition from the wastewater treatment plant to marine waters
- PMID: 38685074
- PMCID: PMC11057073
- DOI: 10.1186/s40793-024-00569-2
Microbial communities colonising plastics during transition from the wastewater treatment plant to marine waters
Abstract
Background: Plastics pollution and antimicrobial resistance (AMR) are two major environmental threats, but potential connections between plastic associated biofilms, the 'plastisphere', and dissemination of AMR genes are not well explored.
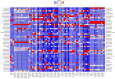
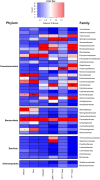
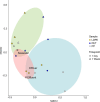
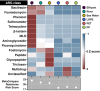
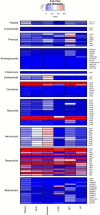
Results: We conducted mesocosm experiments tracking microbial community changes on plastic surfaces transitioning from wastewater effluent to marine environments over 16 weeks. Commonly used plastics, polypropylene (PP), high density polyethylene (HDPE), low density polyethylene (LDPE) and polyethylene terephthalate (PET) incubated in wastewater effluent, river water, estuarine water, and in the seawater for 16 weeks, were analysed via 16S rRNA gene amplicon and shotgun metagenome sequencing. Within one week, plastic-colonizing communities shifted from wastewater effluent-associated microorganisms to marine taxa, some members of which (e.g. Oleibacter-Thalassolituus and Sphingomonas spp., on PET, Alcanivoracaceae on PET and PP, or Oleiphilaceae, on all polymers), were selectively enriched from levels undetectable in the starting communities. Remarkably, microbial biofilms were also susceptible to parasitism, with Saprospiraceae feeding on biofilms at late colonisation stages (from week 6 onwards), while Bdellovibrionaceae were prominently present on HDPE from week 2 and LDPE from day 1. Relative AMR gene abundance declined over time, and plastics did not become enriched for key AMR genes after wastewater exposure.
Conclusion: Although some resistance genes occurred during the mesocosm transition on plastic substrata, those originated from the seawater organisms. Overall, plastic surfaces incubated in wastewater did not act as hotspots for AMR proliferation in simulated marine environments.
© 2024. The Author(s).
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures


References
Grants and funding
- NE/S004548/Natural Environment Research Council
- NE/S004548/Natural Environment Research Council
- NE/S004548/Natural Environment Research Council
- NE/S004548/Natural Environment Research Council
- NE/S004548/Natural Environment Research Council
- NE/S005501/1/Natural Environment Research Council
- NE/S005501/1/Natural Environment Research Council
- NE/S004548/Natural Environment Research Council
- NE/S004548/Natural Environment Research Council
- NE/S004548/Natural Environment Research Council
- 81280/European Regional Development Fund
- 81280/European Regional Development Fund
- 81280/European Regional Development Fund
- 81280/European Regional Development Fund
- 81280/European Regional Development Fund
- 81280/European Regional Development Fund
- 81280/European Regional Development Fund
- 101000327/Horizon 2020 Framework Programme
- 101000327/Horizon 2020 Framework Programme
- 101000327/Horizon 2020 Framework Programme
LinkOut - more resources
Full Text Sources
Miscellaneous
