A novel defined programmed cell death related gene signature for predicting the prognosis of serous ovarian cancer
- PMID: 38685095
- PMCID: PMC11057167
- DOI: 10.1186/s13048-024-01419-y
A novel defined programmed cell death related gene signature for predicting the prognosis of serous ovarian cancer
Abstract
Purpose: This study aims to explore the contribution of differentially expressed programmed cell death genes (DEPCDGs) to the heterogeneity of serous ovarian cancer (SOC) through single-cell RNA sequencing (scRNA-seq) and assess their potential as predictors for clinical prognosis.
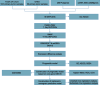
Methods: SOC scRNA-seq data were extracted from the Gene Expression Omnibus database, and the principal component analysis was used for cell clustering. Bulk RNA-seq data were employed to analyze SOC-associated immune cell subsets key genes. CIBERSORT and single-sample gene set enrichment analysis (ssGSEA) were utilized to calculate immune cell scores. Prognostic models and nomograms were developed through univariate and multivariate Cox analyses.
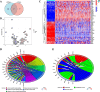
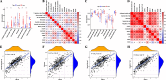
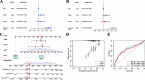
Results: Our analysis revealed that 48 DEPCDGs are significantly correlated with apoptotic signaling and oxidative stress pathways and identified seven key DEPCDGs (CASP3, GADD45B, GNA15, GZMB, IL1B, ISG20, and RHOB) through survival analysis. Furthermore, eight distinct cell subtypes were characterized using scRNA-seq. It was found that G protein subunit alpha 15 (GNA15) exhibited low expression across these subtypes and a strong association with immune cells. Based on the DEGs identified by the GNA15 high- and low-expression groups, a prognostic model comprising eight genes with significant prognostic value was constructed, effectively predicting patient overall survival. Additionally, a nomogram incorporating the RS signature, age, grade, and stage was developed and validated using two large SOC datasets.
Conclusion: GNA15 emerged as an independent and excellent prognostic marker for SOC patients. This study provides valuable insights into the prognostic potential of DEPCDGs in SOC, presenting new avenues for personalized treatment strategies.
Keywords: Clinical prediction; Ovarian cancer; Overall survival; Precision medicine; Serous ovarian cancer; Single-cell RNA sequencing.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

