The TP53-activated E3 ligase RNF144B is a tumour suppressor that prevents genomic instability
- PMID: 38685100
- PMCID: PMC11057071
- DOI: 10.1186/s13046-024-03045-4
The TP53-activated E3 ligase RNF144B is a tumour suppressor that prevents genomic instability
Abstract
Background: TP53, the most frequently mutated gene in human cancers, orchestrates a complex transcriptional program crucial for cancer prevention. While certain TP53-dependent genes have been extensively studied, others, like the recently identified RNF144B, remained poorly understood. This E3 ubiquitin ligase has shown potent tumor suppressor activity in murine Eμ Myc-driven lymphoma, emphasizing its significance in the TP53 network. However, little is known about its targets and its role in cancer development, requiring further exploration. In this work, we investigate RNF144B's impact on tumor suppression beyond the hematopoietic compartment in human cancers.
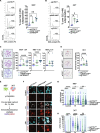
Methods: Employing TP53 wild-type cells, we generated models lacking RNF144B in both non-transformed and cancerous cells of human and mouse origin. By using proteomics, transcriptomics, and functional analysis, we assessed RNF144B's impact in cellular proliferation and transformation. Through in vitro and in vivo experiments, we explored proliferation, DNA repair, cell cycle control, mitotic progression, and treatment resistance. Findings were contrasted with clinical datasets and bioinformatics analysis.
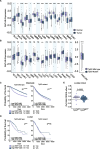
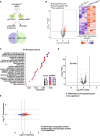
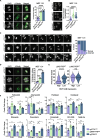
Results: Our research underscores RNF144B's pivotal role as a tumor suppressor, particularly in lung adenocarcinoma. In both human and mouse oncogene-expressing cells, RNF144B deficiency heightened cellular proliferation and transformation. Proteomic and transcriptomic analysis revealed RNF144B's novel function in mediating protein degradation associated with cell cycle progression, DNA damage response and genomic stability. RNF144B deficiency induced chromosomal instability, mitotic defects, and correlated with elevated aneuploidy and worse prognosis in human tumors. Furthermore, RNF144B-deficient lung adenocarcinoma cells exhibited resistance to cell cycle inhibitors that induce chromosomal instability.
Conclusions: Supported by clinical data, our study suggests that RNF144B plays a pivotal role in maintaining genomic stability during tumor suppression.
Keywords: Aneuploidy; Cancer; Genomic instability; Tumor suppressor.
© 2024. The Author(s).
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures

References
-
- Cheok CF, Verma CS, Baselga J, Lane DP. Translating p53 into the clinic. Nat Rev Clin Oncol. 2011;8. 10.1038/nrclinonc.2010.174. - PubMed
-
- McBride KA, Ballinger ML, Killick E, Kirk J, Tattersall MHN, Eeles RA, et al. Li-Fraumeni syndrome: Cancer risk assessment and clinical management. Nat Rev Clin Oncol. 2014;11. 10.1038/nrclinonc.2014.41. - PubMed
-
- Schneider K, Zelley K, Nichols KE, Garber J. Li-Fraumeni Syndrome. 1999. 2019. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

