BitterMasS: Predicting Bitterness from Mass Spectra
- PMID: 38685906
- PMCID: PMC11082931
- DOI: 10.1021/acs.jafc.3c09767
BitterMasS: Predicting Bitterness from Mass Spectra
Abstract
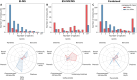
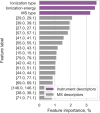
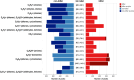
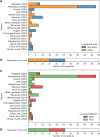
Bitter compounds are common in nature and among drugs. Previously, machine learning tools were developed to predict bitterness from the chemical structure. However, known structures are estimated to represent only 5-10% of the metabolome, and the rest remain unassigned or "dark". We present BitterMasS, a Random Forest classifier that was trained on 5414 experimental mass spectra of bitter and nonbitter compounds, achieving precision = 0.83 and recall = 0.90 for an internal test set. Next, the model was tested against spectra newly extracted from the literature 106 bitter and nonbitter compounds and for additional spectra measured for 26 compounds. For these external test cases, BitterMasS exhibited 67% precision and 93% recall for the first and 58% accuracy and 99% recall for the second. The spectrum-bitterness prediction strategy was more effective than the spectrum-structure-bitterness prediction strategy and covered more compounds. These encouraging results suggest that BitterMasS can be used to predict bitter compounds in the metabolome without the need for structural assignment of individual molecules. This may enable identification of bitter compounds from metabolomics analyses, for comparing potential bitterness levels obtained by different treatments of samples and for monitoring bitterness changes overtime.
Keywords: bitterness; classifier; machine learning; mass spectra; metabolome; natural products; taste.
Conflict of interest statement
The authors declare no competing financial interest.
Figures

Similar articles
-
Research on Bitter Peptides in the Field of Bioinformatics: A Comprehensive Review.Int J Mol Sci. 2024 Sep 12;25(18):9844. doi: 10.3390/ijms25189844. Int J Mol Sci. 2024. PMID: 39337334 Free PMC article. Review.
-
BitterSweetForest: A Random Forest Based Binary Classifier to Predict Bitterness and Sweetness of Chemical Compounds.Front Chem. 2018 Apr 11;6:93. doi: 10.3389/fchem.2018.00093. eCollection 2018. Front Chem. 2018. PMID: 29696137 Free PMC article.
-
CBDPS 1.0: A Python GUI Application for Machine Learning Models to Predict Bitter-Tasting Children's Oral Medicines.Chem Pharm Bull (Tokyo). 2021 Oct 1;69(10):989-994. doi: 10.1248/cpb.c20-00866. Epub 2021 Aug 21. Chem Pharm Bull (Tokyo). 2021. PMID: 34421065
-
Bitter or not? BitterPredict, a tool for predicting taste from chemical structure.Sci Rep. 2017 Sep 21;7(1):12074. doi: 10.1038/s41598-017-12359-7. Sci Rep. 2017. PMID: 28935887 Free PMC article.
-
Sensory evaluation, chemical structures, and threshold concentrations of bitter-tasting compounds in common foodstuffs derived from plants and maillard reaction: A review.Crit Rev Food Sci Nutr. 2023;63(14):2277-2317. doi: 10.1080/10408398.2021.1973956. Epub 2021 Sep 20. Crit Rev Food Sci Nutr. 2023. PMID: 34542344 Review.
Cited by
-
Research on Bitter Peptides in the Field of Bioinformatics: A Comprehensive Review.Int J Mol Sci. 2024 Sep 12;25(18):9844. doi: 10.3390/ijms25189844. Int J Mol Sci. 2024. PMID: 39337334 Free PMC article. Review.
-
BitterDB: 2024 update on bitter ligands and taste receptors.Nucleic Acids Res. 2025 Jan 6;53(D1):D1645-D1650. doi: 10.1093/nar/gkae1044. Nucleic Acids Res. 2025. PMID: 39535052 Free PMC article.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

