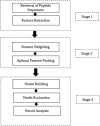
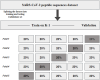
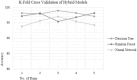
Physicochemical properties-based hybrid machine learning technique for the prediction of SARS-CoV-2 T-cell epitopes as vaccine targets
- PMID: 38686005
- PMCID: PMC11057572
- DOI: 10.7717/peerj-cs.1980
Physicochemical properties-based hybrid machine learning technique for the prediction of SARS-CoV-2 T-cell epitopes as vaccine targets
Abstract
Majority of the existing SARS-CoV-2 vaccines work by presenting the whole pathogen in the attenuated form to immune system to invoke an immune response. On the other hand, the concept of a peptide based vaccine (PBV) is based on the identification and chemical synthesis of only immunodominant peptides known as T-cell epitopes (TCEs) to induce a specific immune response against a particular pathogen. However PBVs have received less attention despite holding huge untapped potential for boosting vaccine safety and immunogenicity. To identify these TCEs for designing PBV, wet-lab experiments are difficult, expensive, and time-consuming. Machine learning (ML) techniques can accurately predict TCEs, saving time and cost for speedy vaccine development. This work proposes novel hybrid ML techniques based on the physicochemical properties of peptides to predict SARS-CoV-2 TCEs. The proposed hybrid ML technique was evaluated using various ML model evaluation metrics and demonstrated promising results. The hybrid technique of decision tree classifier with chi-squared feature weighting technique and forward search optimal feature searching algorithm has been identified as the best model with an accuracy of 98.19%. Furthermore, K-fold cross-validation (KFCV) was performed to ensure that the model is reliable and the results indicate that the hybrid random forest model performs consistently well in terms of accuracy with respect to other hybrid approaches. The predicted TCEs are highly likely to serve as promising vaccine targets, subject to evaluations both in-vivo and in-vitro. This development could potentially save countless lives globally, prevent future epidemic-scale outbreaks, and reduce the risk of mutation escape.
Keywords: COVID-19; Hybrid technique; Machine learning; Peptide based vaccine; SARS-CoV-2; T-cell epitope.
© 2024 Bukhari et al.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures
Similar articles
-
Hybrid Predictive Machine Learning Model for the Prediction of Immunodominant Peptides of Respiratory Syncytial Virus.Bioengineering (Basel). 2024 Aug 5;11(8):791. doi: 10.3390/bioengineering11080791. Bioengineering (Basel). 2024. PMID: 39199749 Free PMC article.
-
Decision tree based ensemble machine learning model for the prediction of Zika virus T-cell epitopes as potential vaccine candidates.Sci Rep. 2022 May 12;12(1):7810. doi: 10.1038/s41598-022-11731-6. Sci Rep. 2022. PMID: 35552469 Free PMC article.
-
Ensemble Machine Learning Model to Predict SARS-CoV-2 T-Cell Epitopes as Potential Vaccine Targets.Diagnostics (Basel). 2021 Oct 26;11(11):1990. doi: 10.3390/diagnostics11111990. Diagnostics (Basel). 2021. PMID: 34829338 Free PMC article.
-
Machine Learning Techniques for the Prediction of B-Cell and T-Cell Epitopes as Potential Vaccine Targets with a Specific Focus on SARS-CoV-2 Pathogen: A Review.Pathogens. 2022 Jan 24;11(2):146. doi: 10.3390/pathogens11020146. Pathogens. 2022. PMID: 35215090 Free PMC article. Review.
-
Development of multi-epitope peptide-based vaccines against SARS-CoV-2.Biomed J. 2021 Mar;44(1):18-30. doi: 10.1016/j.bj.2020.09.005. Epub 2020 Oct 1. Biomed J. 2021. PMID: 33727051 Free PMC article. Review.
Cited by
-
Arabic dialect identification in social media: A hybrid model with transformer models and BiLSTM.Heliyon. 2024 Aug 13;10(17):e36280. doi: 10.1016/j.heliyon.2024.e36280. eCollection 2024 Sep 15. Heliyon. 2024. PMID: 39296033 Free PMC article.
References
-
- Alpaydin E. Introduction to machine learning. Second Edition. Cambridge, MA: The MIT Press; 2010.
-
- Bradley AP. The use of the area under the ROC curve in the evaluation of machine learning algorithms. Pattern Recognition. 1997;30(7):1145–1159. doi: 10.1016/S0031-3203(96)00142-2. - DOI
LinkOut - more resources
Full Text Sources
Miscellaneous