The clinical importance of metagenomic next-generation sequencing in detecting disease-causing microorganisms in cases of sepsis acquired in the community or hospital setting
- PMID: 38686114
- PMCID: PMC11056561
- DOI: 10.3389/fmicb.2024.1384166
The clinical importance of metagenomic next-generation sequencing in detecting disease-causing microorganisms in cases of sepsis acquired in the community or hospital setting
Abstract
Objectives: Although metagenomic next-generation sequencing (mNGS) is commonly used for diagnosing infectious diseases, clinicians face limited options due to the high costs that are not covered by basic medical insurance. The goal of this research is to challenge this bias through a thorough examination and evaluation of the clinical importance of mNGS in precisely identifying pathogenic microorganisms in cases of sepsis acquired in the community or in hospitals.
Methods: A retrospective observational study took place at a tertiary teaching hospital in China from January to December 2021. Data on 308 sepsis patients were collected, and the performance of etiological examination was compared between mNGS and traditional culture method.
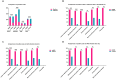
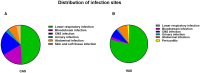
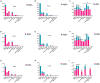
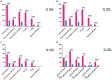
Results: Two hundred twenty-nine cases were observed in the community-acquired sepsis (CAS) group and 79 cases in the hospital-acquired sepsis (HAS) group. In comparison with conventional culture, mNGS showed a significantly higher rate of positivity in both the CAS group (88.21% vs. 25.76%, adj.P < 0.001) and the HAS group (87.34% vs. 44.30%, adj.P < 0.001), particularly across various infection sites and specimens, which were not influenced by factors like antibiotic exposure or the timing and frequency of mNGS technology. Sepsis pathogens detected by mNGS were broad, especially viruses, Mycobacterium tuberculosis, and atypical pathogens, with mixed pathogens being common, particularly bacterial-viral co-detection. Based on the optimization of antimicrobial therapy using mNGS, 58 patients underwent antibiotic de-escalation, two patients were switched to antiviral therapy, and 14 patients initiated treatment for tuberculosis, resulting in a reduction in antibiotic overuse but without significant impact on sepsis prognosis. The HAS group exhibited a critical condition, poor prognosis, high medical expenses, and variations in etiology, yet the mNGS results did not result in increased medical costs for either group.
Conclusions: mNGS demonstrates efficacy in identifying multiple pathogens responsible for sepsis, with mixed pathogens of bacteria and viruses being prevalent. Variability in microbiological profiles among different infection setting underscores the importance of clinical vigilance. Therefore, the adoption of mNGS for microbiological diagnosis of sepsis warrants acknowledgment and promotion.
Keywords: community-acquired sepsis; hospital-acquired sepsis; mNGS; medical expenses; microorganisms; optimizing antimicrobial therapy.
Copyright © 2024 Zhang, Li, Wang, Zhao and Zhang.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
Similar articles
-
Metagenomic Next-Generation Sequencing for the Microbiological Diagnosis of Abdominal Sepsis Patients.Front Microbiol. 2022 Feb 2;13:816631. doi: 10.3389/fmicb.2022.816631. eCollection 2022. Front Microbiol. 2022. PMID: 35185847 Free PMC article.
-
Utility of Metagenomic Next-Generation Sequencing for Etiological Diagnosis of Patients with Sepsis in Intensive Care Units.Microbiol Spectr. 2022 Aug 31;10(4):e0074622. doi: 10.1128/spectrum.00746-22. Epub 2022 Jul 21. Microbiol Spectr. 2022. PMID: 35861525 Free PMC article.
-
The clinical value of metagenomic next-generation sequencing in the microbiological diagnosis of skin and soft tissue infections.Int J Infect Dis. 2020 Nov;100:414-420. doi: 10.1016/j.ijid.2020.09.007. Epub 2020 Sep 6. Int J Infect Dis. 2020. PMID: 32898669
-
Pathogen detection in suspected spinal infection: metagenomic next-generation sequencing versus culture.Eur Spine J. 2023 Dec;32(12):4220-4228. doi: 10.1007/s00586-023-07707-3. Epub 2023 May 26. Eur Spine J. 2023. PMID: 37237239 Review.
-
Rapid diagnosis of Talaromyces marneffei infection by metagenomic next-generation sequencing technology in a Chinese cohort of inborn errors of immunity.Front Cell Infect Microbiol. 2022 Sep 8;12:987692. doi: 10.3389/fcimb.2022.987692. eCollection 2022. Front Cell Infect Microbiol. 2022. PMID: 36159645 Free PMC article.
Cited by
-
An overview of rapid non-culture-based techniques in various clinical specimens for the laboratory diagnosis of Talaromyces marneffei.Front Cell Infect Microbiol. 2025 May 23;15:1591429. doi: 10.3389/fcimb.2025.1591429. eCollection 2025. Front Cell Infect Microbiol. 2025. PMID: 40487313 Free PMC article. Review.
-
Correlation between the gut microbiota characteristics of hosts with severe acute pancreatitis and secondary intra-abdominal infection.Front Med (Lausanne). 2024 Aug 21;11:1409409. doi: 10.3389/fmed.2024.1409409. eCollection 2024. Front Med (Lausanne). 2024. PMID: 39234039 Free PMC article.
References
-
- Adegbite B. R., Elegbede-Adegbite N. O. M., Edoa J. R., Honkpehedji Y. J., Zinsou J. F., Dejon-Agobé J. C., et al. . (2023). Clinical features, treatment outcomes and mortality risk of tuberculosis sepsis in HIV-negative patients: a systematic review and meta-analysis of case reports. Infection 51, 609–621. 10.1007/s15010-022-01950-4 - DOI - PMC - PubMed
-
- Bollinger M., Frère N., Shapeton A. D., Schary W., Kohl M., Kill C., et al. . (2023). Does prehospital suspicion of sepsis shorten time to administration of antibiotics in the emergency department? A retrospective study in one university hospital. J. Clin. Med. 12:5639. 10.3390/jcm12175639 - DOI - PMC - PubMed
-
- Cheng M. P., Stenstrom R., Paquette K., Stabler S. N., Akhter M., Davidson A. C., et al. . (2019). Blood culture results before and after antimicrobial administration in patients with severe manifestations of sepsis: a diagnostic study. Ann. Intern. Med. 171, 547–554. 10.7326/M19-1696 - DOI - PubMed
LinkOut - more resources
Full Text Sources

