Identification and validation of aging-related genes in heart failure based on multiple machine learning algorithms
- PMID: 38686376
- PMCID: PMC11056574
- DOI: 10.3389/fimmu.2024.1367235
Identification and validation of aging-related genes in heart failure based on multiple machine learning algorithms
Abstract
Background: In the face of continued growth in the elderly population, the need to understand and combat age-related cardiac decline becomes even more urgent, requiring us to uncover new pathological and cardioprotective pathways.
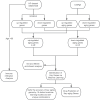
Methods: We obtained the aging-related genes of heart failure through WGCNA and CellAge database. We elucidated the biological functions and signaling pathways involved in heart failure and aging through GO and KEGG enrichment analysis. We used three machine learning algorithms: LASSO, RF and SVM-RFE to further screen the aging-related genes of heart failure, and fitted and verified them through a variety of machine learning algorithms. We searched for drugs to treat age-related heart failure through the DSigDB database. Finally, We use CIBERSORT to complete immune infiltration analysis of aging samples.
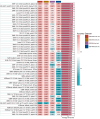
Results: We obtained 57 up-regulated and 195 down-regulated aging-related genes in heart failure through WGCNA and CellAge databases. GO and KEGG enrichment analysis showed that aging-related genes are mainly involved in mechanisms such as Cellular senescence and Cell cycle. We further screened aging-related genes through machine learning and obtained 14 key genes. We verified the results on the test set and 2 external validation sets using 15 machine learning algorithm models and 207 combinations, and the highest accuracy was 0.911. Through screening of the DSigDB database, we believe that rimonabant and lovastatin have the potential to delay aging and protect the heart. The results of immune infiltration analysis showed that there were significant differences between Macrophages M2 and T cells CD8 in aging myocardium.
Conclusion: We identified aging signature genes and potential therapeutic drugs for heart failure through bioinformatics and multiple machine learning algorithms, providing new ideas for studying the mechanism and treatment of age-related cardiac decline.
Keywords: aging; bioinformatics; heart failure; immune infiltration analysis; machine learning.
Copyright © 2024 Yu, Wang, Hou, Xue, Liu and Li.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




Similar articles
-
Decoding the cytokine code for heart failure based on bioinformatics, machine learning and Bayesian networks.Biochim Biophys Acta Mol Basis Dis. 2025 Mar;1871(3):167701. doi: 10.1016/j.bbadis.2025.167701. Epub 2025 Feb 4. Biochim Biophys Acta Mol Basis Dis. 2025. PMID: 39909085
-
Exploration of the shared diagnostic genes and mechanisms between periodontitis and primary Sjögren's syndrome by integrated comprehensive bioinformatics analysis and machine learning.Int Immunopharmacol. 2024 Nov 15;141:112899. doi: 10.1016/j.intimp.2024.112899. Epub 2024 Aug 13. Int Immunopharmacol. 2024. PMID: 39142001
-
Screening of key immune-related gene in Parkinson's disease based on WGCNA and machine learning.Zhong Nan Da Xue Xue Bao Yi Xue Ban. 2024 Feb 28;49(2):207-219. doi: 10.11817/j.issn.1672-7347.2024.230307. Zhong Nan Da Xue Xue Bao Yi Xue Ban. 2024. PMID: 38755717 Free PMC article. Chinese, English.
-
Identification and Verification of the Oxidative Stress-Related Signature Markers for Intracranial Aneurysm-Applied Bioinformatics.Front Biosci (Landmark Ed). 2024 Aug 20;29(8):294. doi: 10.31083/j.fbl2908294. Front Biosci (Landmark Ed). 2024. PMID: 39206894
-
Bioinformatics and machine learning approaches reveal key genes and underlying molecular mechanisms of atherosclerosis: A review.Medicine (Baltimore). 2024 Aug 2;103(31):e38744. doi: 10.1097/MD.0000000000038744. Medicine (Baltimore). 2024. PMID: 39093811 Free PMC article. Review.
Cited by
-
Commentary: Immune cell infiltration and prognostic index in cervical cancer: insights from metabolism-related differential genes.Front Immunol. 2024 Sep 19;15:1446741. doi: 10.3389/fimmu.2024.1446741. eCollection 2024. Front Immunol. 2024. PMID: 39364407 Free PMC article. No abstract available.
-
Plin2 Coordinates Immune and Metabolic Reprogramming in Lacrimal Gland Aging.Invest Ophthalmol Vis Sci. 2025 Jun 2;66(6):79. doi: 10.1167/iovs.66.6.79. Invest Ophthalmol Vis Sci. 2025. PMID: 40569117 Free PMC article.
References
-
- Savarese G, Becher PM, Lund LH, Seferovic P, Rosano GMC, Coats AJS. Global burden of heart failure: a comprehensive and updated review of epidemiology [published correction appears in Cardiovasc Res. 2023 Jun 13;119(6):1453]. Cardiovasc Res. (2023) 118:3272–87. doi: 10.1093/cvr/cvac013 - DOI - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Research Materials

